Deciphering RNA G-quadruplex function during the early steps of HIV-1 infection
- PMID: 36453997
- PMCID: PMC9757044
- DOI: 10.1093/nar/gkac1030
Deciphering RNA G-quadruplex function during the early steps of HIV-1 infection
Abstract
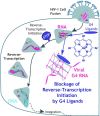
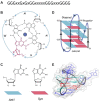
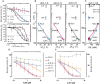
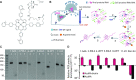
G-quadruplexes (G4s) are four-stranded nucleic acid structures formed by the stacking of G-tetrads. Here we investigated their formation and function during HIV-1 infection. Using bioinformatics and biophysics analyses we first searched for evolutionary conserved G4-forming sequences in HIV-1 genome. We identified 10 G4s with conservation rates higher than those of HIV-1 regulatory sequences such as RRE and TAR. We then used porphyrin-based G4-binders to probe the formation of the G4s during infection of human cells by native HIV-1. The G4-binders efficiently inhibited HIV-1 infectivity, which is attributed to the formation of G4 structures during HIV-1 replication. Using a qRT-PCR approach, we showed that the formation of viral G4s occurs during the first 2 h post-infection and their stabilization by the G4-binders prevents initiation of reverse transcription. We also used a G4-RNA pull-down approach, based on a G4-specific biotinylated probe, to allow the direct detection and identification of viral G4-RNA in infected cells. Most of the detected G4-RNAs contain crucial regulatory elements such as the PPT and cPPT sequences as well as the U3 region. Hence, these G4s would function in the early stages of infection when the viral RNA genome is being processed for the reverse transcription step.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Paeschke K., Simonsson T., Postberg J., Rhodes D., Lipps H.J.. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005; 12:847–854. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

