Growth condition-dependent differences in methylation imply transiently differentiated DNA methylation states in Escherichia coli
- PMID: 36454087
- PMCID: PMC9911048
- DOI: 10.1093/g3journal/jkac310
Growth condition-dependent differences in methylation imply transiently differentiated DNA methylation states in Escherichia coli
Abstract
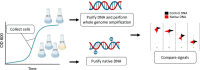
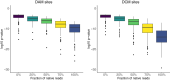
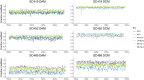
DNA methylation in bacteria frequently serves as a simple immune system, allowing recognition of DNA from foreign sources, such as phages or selfish genetic elements. However, DNA methylation also affects other cell phenotypes in a heritable manner (i.e. epigenetically). While there are several examples of methylation affecting transcription in an epigenetic manner in highly localized contexts, it is not well-established how frequently methylation serves a more general epigenetic function over larger genomic scales. To address this question, here we use Oxford Nanopore sequencing to profile DNA modification marks in three natural isolates of Escherichia coli. We first identify the DNA sequence motifs targeted by the methyltransferases in each strain. We then quantify the frequency of methylation at each of these motifs across the entire genome in different growth conditions. We find that motifs in specific regions of the genome consistently exhibit high or low levels of methylation. Furthermore, we show that there are replicable and consistent differences in methylated regions across different growth conditions. This suggests that during growth, E. coli transiently differentiate into distinct methylation states that depend on the growth state, raising the possibility that measuring DNA methylation alone can be used to infer bacterial growth states without additional information such as transcriptome or proteome data. These results show the utility of using Oxford Nanopore sequencing as an economic means to infer DNA methylation status. They also provide new insights into the dynamics of methylation during bacterial growth and provide evidence of differentiated cell states, a transient analog to what is observed in the differentiation of cell types in multicellular organisms.
Keywords: E coli; DNA methylation; nanopore.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Genetics Society of America.
Conflict of interest statement
Conflict of interest None declared.
Figures


References
-
- Adzitey F, Asante J, Kumalo HM, Khan RB, Somboro AM, Amoako DG. Genomic investigation into the virulome, pathogenicity, stress response factors, clonal lineages, and phylogenetic relationship of Escherichia coli strains isolated from meat sources in Ghana. Genes (Basel). 2020;11(12):12. doi: 10.3390/genes11121504. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
