LambdaPP: Fast and accessible protein-specific phenotype predictions
- PMID: 36454227
- PMCID: PMC9793974
- DOI: 10.1002/pro.4524
LambdaPP: Fast and accessible protein-specific phenotype predictions
Abstract
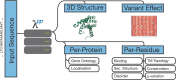
The availability of accurate and fast artificial intelligence (AI) solutions predicting aspects of proteins are revolutionizing experimental and computational molecular biology. The webserver LambdaPP aspires to supersede PredictProtein, the first internet server making AI protein predictions available in 1992. Given a protein sequence as input, LambdaPP provides easily accessible visualizations of protein 3D structure, along with predictions at the protein level (GeneOntology, subcellular location), and the residue level (binding to metal ions, small molecules, and nucleotides; conservation; intrinsic disorder; secondary structure; alpha-helical and beta-barrel transmembrane segments; signal-peptides; variant effect) in seconds. The structure prediction provided by LambdaPP-leveraging ColabFold and computed in minutes-is based on MMseqs2 multiple sequence alignments. All other feature prediction methods are based on the pLM ProtT5. Queried by a protein sequence, LambdaPP computes protein and residue predictions almost instantly for various phenotypes, including 3D structure and aspects of protein function. LambdaPP is freely available for everyone to use under embed.predictprotein.org, the interactive results for the case study can be found under https://embed.predictprotein.org/o/Q9NZC2. The frontend of LambdaPP can be found on GitHub (github.com/sacdallago/embed.predictprotein.org), and can be freely used and distributed under the academic free use license (AFL-2). For high-throughput applications, all methods can be executed locally via the bio-embeddings (bioembeddings.com) python package, or docker image at ghcr.io/bioembeddings/bio_embeddings, which also includes the backend of LambdaPP.
Keywords: artificial intelligence; protein annotation; protein function prediction; protein language models; protein structure prediction; web server.
© 2022 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Abriata LA, Tamò GE, Monastyrskyy B, Kryshtafovych A, Dal Peraro M. Assessment of hard target modeling in CASP12 reveals an emerging role of alignment‐based contact prediction methods. Proteins. 2018;86(Suppl. 1):97–112. - PubMed
-
- Ahdritz G, Bouatta N, Kadyan S, Xia Q, Gerecke W, AlQuraishi M. OpenFold. 2021.
-
- Alexander‐Brett JM, Kober DL. Triggering receptor expressed on myeloid cells 2. 2015. 10.2210/pdb5ELI/pdb - DOI
-
- Almagro Armenteros JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33:3387–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

