Wheat EARLY FLOWERING 3 affects heading date without disrupting circadian oscillations
- PMID: 36454669
- PMCID: PMC9922389
- DOI: 10.1093/plphys/kiac544
Wheat EARLY FLOWERING 3 affects heading date without disrupting circadian oscillations
Abstract
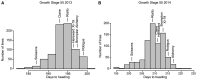
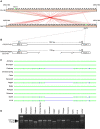
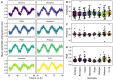
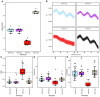
Plant breeders have indirectly selected for variation at circadian-associated loci in many of the world's major crops, when breeding to increase yield and improve crop performance. Using an eight-parent Multiparent Advanced Generation Inter-Cross (MAGIC) population, we investigated how variation in circadian clock-associated genes contributes to the regulation of heading date in UK and European winter wheat (Triticum aestivum) varieties. We identified homoeologues of EARLY FLOWERING 3 (ELF3) as candidates for the Earliness per se (Eps) D1 and B1 loci under field conditions. We then confirmed a single-nucleotide polymorphism within the coding region of TaELF3-B1 as a candidate polymorphism underlying the Eps-B1 locus. We found that a reported deletion at the Eps-D1 locus encompassing TaELF3-D1 is, instead, an allele that lies within an introgression region containing an inversion relative to the Chinese Spring D genome. Using Triticum turgidum cv. Kronos carrying loss-of-function alleles of TtELF3, we showed that ELF3 regulates heading, with loss of a single ELF3 homoeologue sufficient to alter heading date. These studies demonstrated that ELF3 forms part of the circadian oscillator; however, the loss of all homoeologues was required to affect circadian rhythms. Similarly, loss of functional LUX ARRHYTHMO (LUX) in T. aestivum, an orthologue of a protein partner of Arabidopsis (Arabidopsis thaliana) ELF3, severely disrupted circadian rhythms. ELF3 and LUX transcripts are not co-expressed at dusk, suggesting that the structure of the wheat circadian oscillator might differ from that of Arabidopsis. Our demonstration that alterations to ELF3 homoeologues can affect heading date separately from effects on the circadian oscillator suggests a role for ELF3 in cereal photoperiodic responses that could be selected for without pleiotropic deleterious alterations to circadian rhythms.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



Similar articles
-
Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum).J Exp Bot. 2016 Jan;67(1):287-99. doi: 10.1093/jxb/erv458. Epub 2015 Oct 17. J Exp Bot. 2016. PMID: 26476691 Free PMC article.
-
Genetic and physical mapping of the earliness per se locus Eps-A (m) 1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene.Funct Integr Genomics. 2016 Jul;16(4):365-82. doi: 10.1007/s10142-016-0490-3. Epub 2016 Apr 16. Funct Integr Genomics. 2016. PMID: 27085709 Free PMC article.
-
EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock.Plant Cell. 2012 Feb;24(2):428-43. doi: 10.1105/tpc.111.093807. Epub 2012 Feb 10. Plant Cell. 2012. PMID: 22327739 Free PMC article.
-
Revisiting the role and mechanism of ELF3 in circadian clock modulation.Gene. 2024 Jun 30;913:148378. doi: 10.1016/j.gene.2024.148378. Epub 2024 Mar 13. Gene. 2024. PMID: 38490512 Review.
-
Molecular and functional dissection of EARLY-FLOWERING 3 (ELF3) and ELF4 in Arabidopsis.Plant Sci. 2021 Feb;303:110786. doi: 10.1016/j.plantsci.2020.110786. Epub 2020 Dec 3. Plant Sci. 2021. PMID: 33487361 Review.
Cited by
-
Compartmentation of photosynthesis gene expression in C4 maize depends on time of day.Plant Physiol. 2023 Nov 22;193(4):2306-2320. doi: 10.1093/plphys/kiad447. Plant Physiol. 2023. PMID: 37555432 Free PMC article.
-
Natural variations of wheat EARLY FLOWERING 3 highlight their contributions to local adaptation through fine-tuning of heading time.Theor Appl Genet. 2023 May 26;136(6):139. doi: 10.1007/s00122-023-04386-y. Theor Appl Genet. 2023. PMID: 37233781
-
Exploring the multifaceted dynamics of flowering time regulation in field crops: Insight and intervention approaches.Plant Genome. 2025 Jun;18(2):e70017. doi: 10.1002/tpg2.70017. Plant Genome. 2025. PMID: 40164968 Free PMC article. Review.
-
Phytochromes transmit photoperiod information via the evening complex in Brachypodium.Genome Biol. 2023 Nov 7;24(1):256. doi: 10.1186/s13059-023-03082-w. Genome Biol. 2023. PMID: 37936225 Free PMC article.
-
Introgressions lead to reference bias in wheat RNA-seq analysis.BMC Biol. 2024 Mar 7;22(1):56. doi: 10.1186/s12915-024-01853-w. BMC Biol. 2024. PMID: 38454464 Free PMC article.
References
-
- Aguilar M, Prieto P (2020) Sequence analysis of wheat subtelomeres reveals a high polymorphism among homoeologous chromosomes. Plant Genome 13(3): e20065. - PubMed
-
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L). Theor Appl Genet 115(5): 721–733 - PubMed
-
- Bentley AR, Horsnell R, Werner CP, Turner AS, Rose GA, Bedard C, Howell P, Wilhelm EP, Mackay IJ, Howells RM, et al. (2013) Short, natural, and extended photoperiod response in BC2F4 lines of bread wheat with different Photoperiod-1 (Ppd-1) alleles. J Exp Bot 64(7): 1783–1793 - PubMed
Publication types
MeSH terms
Grants and funding
- BB/K011790/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M015416/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011194/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S006370/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S002251/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

