Plausible pathway for a host-parasite molecular replication network to increase its complexity through Darwinian evolution
- PMID: 36454734
- PMCID: PMC9714742
- DOI: 10.1371/journal.pcbi.1010709
Plausible pathway for a host-parasite molecular replication network to increase its complexity through Darwinian evolution
Abstract
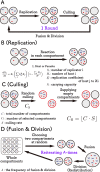
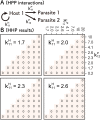
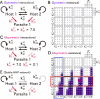
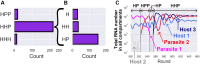
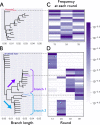
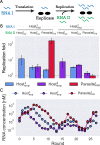
How the complexity of primitive self-replication molecules develops through Darwinian evolution remains a mystery with regards to the origin of life. Theoretical studies have proposed that coevolution with parasitic replicators increases network complexity by inducing inter-dependent replication. Particularly, Takeuchi and Hogeweg proposed a complexification process of replicator networks by successive appearance of a parasitic replicator followed by the addition of a new host replicator that is resistant to the parasitic replicator. However, the feasibility of such complexification with biologically relevant molecules is still unknown owing to the lack of an experimental model. Here, we investigated the plausible complexification pathway of host-parasite replicators using both an experimental host-parasite RNA replication system and a theoretical model based on the experimental system. We first analyzed the parameter space that allows for sustainable replication in various replication networks ranging from a single molecule to three-member networks using computer simulation. The analysis shows that the most plausible complexification pathway from a single host replicator is the addition of a parasitic replicator, followed by the addition of a new host replicator that is resistant to the parasite, consistent with the previous study by Takeuchi and Hogeweg. We also provide evidence that the pathway actually occurred in our previous evolutionary experiment. These results provide experimental evidence that a population of a single replicator spontaneously evolves into multi-replicator networks through coevolution with parasitic replicators.
Copyright: © 2022 Kamiura et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
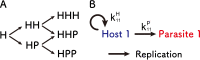


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

