Evaluation of co-circulating pathogens and microbiome from COVID-19 infections
- PMID: 36455065
- PMCID: PMC9714956
- DOI: 10.1371/journal.pone.0278543
Evaluation of co-circulating pathogens and microbiome from COVID-19 infections
Abstract
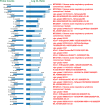
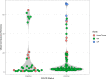
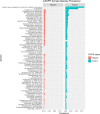
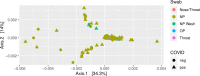
Co-infections or secondary infections with SARS-CoV-2 have the potential to affect disease severity and morbidity. Additionally, the potential influence of the nasal microbiome on COVID-19 illness is not well understood. In this study, we analyzed 203 residual samples, originally submitted for SARS-CoV-2 testing, for the presence of viral, bacterial, and fungal pathogens and non-pathogens using a comprehensive microarray technology, the Lawrence Livermore Microbial Detection Array (LLMDA). Eighty-seven percent of the samples were nasopharyngeal samples, and 23% of the samples were oral, nasal and oral pharyngeal swabs. We conducted bioinformatics analyses to examine differences in microbial populations of these samples, as a proxy for the nasal and oral microbiome, from SARS-CoV-2 positive and negative specimens. We found 91% concordance with the LLMDA relative to a diagnostic RT-qPCR assay for detection of SARS-CoV-2. Sixteen percent of all the samples (32/203) revealed the presence of an opportunistic bacterial or frank viral pathogen with the potential to cause co-infections. The two most detected bacteria, Streptococcus pyogenes and Streptococcus pneumoniae, were present in both SARS-CoV-2 positive and negative samples. Human metapneumovirus was the most prevalent viral pathogen in the SARS-CoV-2 negative samples. Sequence analysis of 16S rRNA was also conducted to evaluate bacterial diversity and confirm LLMDA results.
Copyright: © 2022 Thissen et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

