An integrated resource for functional and structural connectivity of the marmoset brain
- PMID: 36456558
- PMCID: PMC9715556
- DOI: 10.1038/s41467-022-35197-2
An integrated resource for functional and structural connectivity of the marmoset brain
Abstract
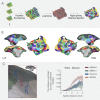
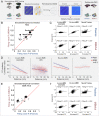
Comprehensive integration of structural and functional connectivity data is required to model brain functions accurately. While resources for studying the structural connectivity of non-human primate brains already exist, their integration with functional connectivity data has remained unavailable. Here we present a comprehensive resource that integrates the most extensive awake marmoset resting-state fMRI data available to date (39 marmoset monkeys, 710 runs, 12117 mins) with previously published cellular-level neuronal tracing data (52 marmoset monkeys, 143 injections) and multi-resolution diffusion MRI datasets. The combination of these data allowed us to (1) map the fine-detailed functional brain networks and cortical parcellations, (2) develop a deep-learning-based parcellation generator that preserves the topographical organization of functional connectivity and reflects individual variabilities, and (3) investigate the structural basis underlying functional connectivity by computational modeling. This resource will enable modeling structure-function relationships and facilitate future comparative and translational studies of primate brains.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

