ENTAIL: yEt aNoTher amyloid fIbrils cLassifier
- PMID: 36456900
- PMCID: PMC9714056
- DOI: 10.1186/s12859-022-05070-6
ENTAIL: yEt aNoTher amyloid fIbrils cLassifier
Abstract
Background: This research aims to increase our knowledge of amyloidoses. These disorders cause incorrect protein folding, affecting protein functionality (on structure). Fibrillar deposits are the basis of some wellknown diseases, such as Alzheimer, Creutzfeldt-Jakob diseases and type II diabetes. For many of these amyloid proteins, the relative precursors are known. Discovering new protein precursors involved in forming amyloid fibril deposits would improve understanding the pathological processes of amyloidoses.
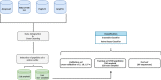
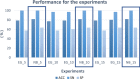
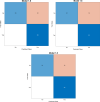
Results: A new classifier, called ENTAIL, was developed using over than 4000 molecular descriptors. ENTAIL was based on the Naive Bayes Classifier with Unbounded Support and Gaussian Kernel Type, with an accuracy on the test set of 81.80%, SN of 100%, SP of 63.63% and an MCC of 0.683 on a balanced dataset.
Conclusions: The analysis carried out has demonstrated how, despite the various configurations of the tests, performances are superior in terms of performance on a balanced dataset.
Keywords: Amyloidoses; Fibrils machine learning; Protein classification.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Citarella AA, Marco FD, Biasi LD, Risi M, Tortora G. Gene ontology terms visualization with dynamic distance-graph and similarity measures (S). In: Chang S, editor. The 27th international DMS conference on visualization and visual languages, DMSVIVA 2021, KSIR Virtual Conference Center, USA, 2021, KSI Research Inc.; 2021. pp. 85–91. 10.18293/DMSVIVA21-013
-
- Citarella AA, Marco FD, Biasi LD, Risi M, Tortora G. PADD: dynamic distance-graph based on similarity measures for GO terms visualization of Alzheimer and Parkinson diseases. J Vis Lang Comput. 2021;2021(1):19–28. doi: 10.18293/JVLC2021-N1-013. - DOI
-
- Allen G. Sequencing of proteins and peptides. Work TS, Burdon R, editors (1981)
-
- Citarella AA, Porcelli L, Di Biasi L, Risi M, Tortora G. Reconstruction and visualization of protein structures by exploiting bidirectional neural networks and discrete classes. In: 2021 25th international conference information visualisation (IV), 2021. pp. 285–290. 10.1109/IV53921.2021.00053. IEEE
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

