Multiplex CRISPR/Cas9-mediated raffinose synthase gene editing reduces raffinose family oligosaccharides in soybean
- PMID: 36457532
- PMCID: PMC9706108
- DOI: 10.3389/fpls.2022.1048967
Multiplex CRISPR/Cas9-mediated raffinose synthase gene editing reduces raffinose family oligosaccharides in soybean
Abstract
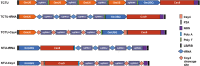
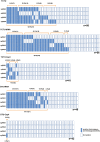
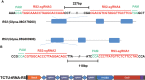
Soybean [Glycine max (L.) Merr.] is an important world economic crop. It is rich in oil, protein, and starch, and soluble carbohydrates in soybean seeds are also important for human and livestock consumption. The predominant soluble carbohydrate in soybean seed is composed of sucrose and raffinose family oligosaccharides (RFOs). Among these carbohydrates, only sucrose can be digested by humans and monogastric animals and is beneficial for metabolizable energy, while RFOs are anti-nutritional factors in diets, usually leading to flatulence and indigestion, ultimately reducing energy efficiency. Hence, breeding efforts to remove RFOs from soybean seeds can increase metabolizable energy and improve nutritional quality. The objective of this research is to use the multiplex Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9-mediated gene editing system to induce the knockout of soybean raffinose synthase (RS) genes RS2 and RS3 simultaneously to reduce RFOs in mature seeds. First, we constructed five types of multiplex gene editing systems and compared their editing efficiency in soybean hairy roots. We confirmed that the two-component transcriptional unit (TCTU) and single transcriptional unit (STU) systems with transfer RNA (tRNA) as the cleavage site performed better than other systems. The average editing efficiency at the four targets with TCTU-tRNA and STU-tRNA was 50.5% and 46.7%, respectively. Then, we designed four single-guide RNA (sgRNA) targets to induce mutations at RS2 and RS3 by using the TCTU-tRNA system. After the soybean transformation, we obtained several RS2 and RS3 mutation plants, and a subset of alleles was successfully transferred to the progeny. We identified null single and double mutants at the T2 generation and analyzed the seed carbohydrate content of their progeny. The RS2 and RS3 double mutants and the RS2 single mutant exhibited dramatically reduced levels of raffinose and stachyose in mature seeds. Further analysis of the growth and development of these mutants showed that there were no penalties on these phenotypes. Our results indicate that knocking out RS genes by multiplex CRISPR/Cas9-mediated gene editing is an efficient way to reduce RFOs in soybean. This research demonstrates the potential of using elite soybean cultivars to improve the soybean meal trait by multiplex CRISPR(Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9-mediated gene editing.
Keywords: CRISPR/Cas9; RFOs; RS; multiplex gene editing; soybean.
Copyright © 2022 Cao, Wang, Ma, Liu, Ji and Duan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

