SRSF1 acts as an IFN-I-regulated cellular dependency factor decisively affecting HIV-1 post-integration steps
- PMID: 36458014
- PMCID: PMC9706209
- DOI: 10.3389/fimmu.2022.935800
SRSF1 acts as an IFN-I-regulated cellular dependency factor decisively affecting HIV-1 post-integration steps
Abstract
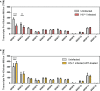
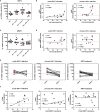
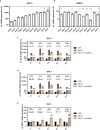
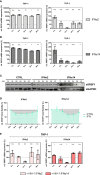
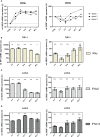
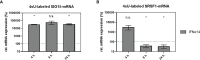
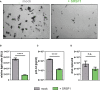
Efficient HIV-1 replication depends on balanced levels of host cell components including cellular splicing factors as the family of serine/arginine-rich splicing factors (SRSF, 1-10). Type I interferons (IFN-I) play a crucial role in the innate immunity against HIV-1 by inducing the expression of IFN-stimulated genes (ISGs) including potent host restriction factors. The less well known IFN-repressed genes (IRepGs) might additionally affect viral replication by downregulating host dependency factors that are essential for the viral life cycle; however, so far, the knowledge about IRepGs involved in HIV-1 infection is very limited. In this work, we could demonstrate that HIV-1 infection and the associated ISG induction correlated with low SRSF1 levels in intestinal lamina propria mononuclear cells (LPMCs) and peripheral blood mononuclear cells (PBMCs) during acute and chronic HIV-1 infection. In HIV-1-susceptible cell lines as well as primary monocyte-derived macrophages (MDMs), expression levels of SRSF1 were transiently repressed upon treatment with specific IFNα subtypes in vitro. Mechanically, 4sU labeling of newly transcribed mRNAs revealed that IFN-mediated SRSF1 repression is regulated on early RNA level. SRSF1 knockdown led to an increase in total viral RNA levels, but the relative proportion of the HIV-1 viral infectivity factor (Vif) coding transcripts, which is essential to counteract APOBEC3G-mediated host restriction, was significantly reduced. In the presence of high APOBEC3G levels, however, increased LTR activity upon SRSF1 knockdown facilitated the overall replication, despite decreased vif mRNA levels. In contrast, SRSF1 overexpression significantly impaired HIV-1 post-integration steps including LTR transcription, alternative splice site usage, and virus particle production. Since balanced SRSF1 levels are crucial for efficient viral replication, our data highlight the so far undescribed role of SRSF1 acting as an IFN-modulated cellular dependency factor decisively regulating HIV-1 post-integration steps.
Keywords: HIV-1; ISG (interferon stimulated genes); SF2/ASF; SRSF1; alternative splicing; interferon; repressed genes; transcription.
Copyright © 2022 Sertznig, Roesmann, Wilhelm, Heininger, Bleekmann, Elsner, Santiago, Schuhenn, Karakoese, Benatzy, Snodgrass, Esser, Sutter, Dittmer and Widera.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

