Modeling interaction networks between host, diet, and bacteria predicts obesogenesis in a mouse model
- PMID: 36458093
- PMCID: PMC9705962
- DOI: 10.3389/fmolb.2022.1059094
Modeling interaction networks between host, diet, and bacteria predicts obesogenesis in a mouse model
Abstract
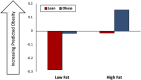
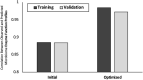
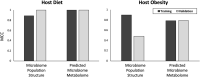
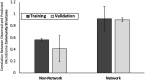
Host-microbiome interactions are known to have substantial effects on human health, but the diversity of the human microbiome makes it difficult to definitively attribute specific microbiome features to a host phenotype. One approach to overcoming this challenge is to use animal models of host-microbiome interaction, but it must be determined that relevant aspects of host-microbiome interactions are reflected in the animal model. One such experimental validation is an experiment by Ridura et al. In that experiment, transplanting a microbiome from a human into a mouse also conferred the human donor's obesity phenotype. We have aggregated a collection of previously published host-microbiome mouse-model experiments and combined it with thousands of sequenced and annotated bacterial genomes and metametabolomic pathways. Three computational models were generated, each model reflecting an aspect of host-microbiome interactions: 1) Predict the change in microbiome community structure in response to host diet using a community interaction network, 2) Predict metagenomic data from microbiome community structure, and 3) Predict host obesogenesis from modeled microbiome metagenomic data. These computationally validated models were combined into an integrated model of host-microbiome-diet interactions and used to replicate the Ridura experiment in silico. The results of the computational models indicate that network-based models are significantly more predictive than similar but non-network-based models. Network-based models also provide additional insight into the molecular mechanisms of host-microbiome interaction by highlighting metabolites and metabolic pathways proposed to be associated with microbiome-based obesogenesis. While the models generated in this study are likely too specific to the animal models and experimental conditions used to train our models to be of general utility in a broader understanding of obesogenesis, the approach detailed here is expected to be a powerful tool of investigating multiple types of host-microbiome interactions.
Keywords: computational modeling; metabolome; microbiome; mouse model; network biology; obesity.
Copyright © 2022 Larsen and Dai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Metabolome of human gut microbiome is predictive of host dysbiosis.Gigascience. 2015 Sep 14;4:42. doi: 10.1186/s13742-015-0084-3. eCollection 2015. Gigascience. 2015. PMID: 26380076 Free PMC article.
-
MiMeNet: Exploring microbiome-metabolome relationships using neural networks.PLoS Comput Biol. 2021 May 17;17(5):e1009021. doi: 10.1371/journal.pcbi.1009021. eCollection 2021 May. PLoS Comput Biol. 2021. PMID: 33999922 Free PMC article.
-
Systematic prediction of health-relevant human-microbial co-metabolism through a computational framework.Gut Microbes. 2015;6(2):120-30. doi: 10.1080/19490976.2015.1023494. Gut Microbes. 2015. PMID: 25901891 Free PMC article.
-
Mapping the inner workings of the microbiome: genomic- and metagenomic-based study of metabolism and metabolic interactions in the human microbiome.Cell Metab. 2014 Nov 4;20(5):742-752. doi: 10.1016/j.cmet.2014.07.021. Epub 2014 Aug 28. Cell Metab. 2014. PMID: 25176148 Free PMC article. Review.
-
Connect the dots: sketching out microbiome interactions through networking approaches.Microbiome Res Rep. 2023 Jul 18;2(4):25. doi: 10.20517/mrr.2023.25. eCollection 2023. Microbiome Res Rep. 2023. PMID: 38058764 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

