Microbial communities in the liver and brain are informative for postmortem submersion interval estimation in the late phase of decomposition: A study in mouse cadavers recovered from freshwater
- PMID: 36458191
- PMCID: PMC9705336
- DOI: 10.3389/fmicb.2022.1052808
Microbial communities in the liver and brain are informative for postmortem submersion interval estimation in the late phase of decomposition: A study in mouse cadavers recovered from freshwater
Abstract
Introduction: Bodies recovered from water, especially in the late phase of decomposition, pose difficulties to the investigating authorities. Various methods have been proposed for postmortem submersion interval (PMSI) estimation and drowning identification, but some limitations remain. Many recent studies have proved the value of microbiota succession in viscera for postmortem interval estimation. Nevertheless, the visceral microbiota succession and its application for PMSI estimation and drowning identification require further investigation.
Methods: In the current study, mouse drowning and CO2 asphyxia models were developed, and cadavers were immersed in freshwater for 0 to 14 days. Microbial communities in the liver and brain were characterized via 16S rDNA high-throughput sequencing.
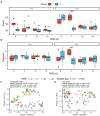
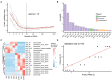
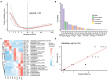
Results: Only livers and brains collected from 5 to 14 days postmortem were qualified for sequencing. There was significant variation between microbiota from liver and brain. Differences in microbiota between the cadavers of mice that had drowned and those only subjected to postmortem submersion decreased over the PMSI. Significant successions in microbial communities were observed among the different subgroups within the late phase of the PMSI in livers and brains. Eighteen taxa in the liver which were mainly related to Clostridium_sensu_stricto and Aeromonas, and 26 taxa in the brain which were mainly belonged to Clostridium_sensu_stricto, Acetobacteroides, and Limnochorda, were selected as potential biomarkers for PMSI estimation based on a random forest algorithm. The PMSI estimation models established yielded accurate prediction results with mean absolute errors ± the standard error of 1.282 ± 0.189 d for the liver and 0.989 ± 0.237 d for the brain.
Conclusions: The present study provides novel information on visceral postmortem microbiota succession in corpses submerged in freshwater which sheds new light on PMSI estimation based on the liver and brain in forensic practice.
Keywords: aquatic habitat; decomposition; internal organ; microbial community; postmortem submersion interval.
Copyright © 2022 Wang, Zhang, Zeng, Dong, Yuan, Wang, Liu, Pan, Zhao and Guan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Postmortem submersion interval estimation of cadavers recovered from freshwater based on gut microbial community succession.Front Microbiol. 2022 Dec 2;13:988297. doi: 10.3389/fmicb.2022.988297. eCollection 2022. Front Microbiol. 2022. PMID: 36532467 Free PMC article.
-
Microbiota signature of the lung as the promising bioindicator for drowning diagnosis and postmortem submersion interval estimation.Int J Legal Med. 2025 Jul;139(4):1863-1877. doi: 10.1007/s00414-025-03458-6. Epub 2025 Mar 11. Int J Legal Med. 2025. PMID: 40067359
-
A fundamental study on postmortem submersion interval estimation by metabolomics analyzing of gastrocnemius muscle from submersed rat models in freshwater.Int J Legal Med. 2024 Sep;138(5):2037-2047. doi: 10.1007/s00414-024-03258-4. Epub 2024 May 28. Int J Legal Med. 2024. PMID: 38802694
-
Research Progress of Aquatic Corpse Decomposition and Postmortem Submersion Interval Estimation.Fa Yi Xue Za Zhi. 2019 Aug;35(4):459-466. doi: 10.12116/j.issn.1004-5619.2019.04.016. Epub 2019 Aug 25. Fa Yi Xue Za Zhi. 2019. PMID: 31532158 Review. Chinese, English.
-
Review and Prospect of Diagnosis of Drowning Deaths in Water.Fa Yi Xue Za Zhi. 2022 Feb 25;38(1):3-13. doi: 10.12116/j.issn.1004-5619.2021.410625. Fa Yi Xue Za Zhi. 2022. PMID: 35725698 Review. Chinese, English.
Cited by
-
Novel thermophilic genera Geochorda gen. nov. and Carboxydochorda gen. nov. from the deep terrestrial subsurface reveal the ecophysiological diversity in the class Limnochordia.Front Microbiol. 2024 Sep 23;15:1441865. doi: 10.3389/fmicb.2024.1441865. eCollection 2024. Front Microbiol. 2024. PMID: 39376703 Free PMC article.
-
Microbial community profiling for forensic drowning diagnosis across locations and submersion times.BMC Microbiol. 2025 Apr 24;25(1):244. doi: 10.1186/s12866-025-03902-y. BMC Microbiol. 2025. PMID: 40275149 Free PMC article.
-
Changes in Microbial Communities Using Pigs as a Model for Postmortem Interval Estimation.Microorganisms. 2023 Nov 20;11(11):2811. doi: 10.3390/microorganisms11112811. Microorganisms. 2023. PMID: 38004822 Free PMC article.
References
-
- Amrani A., Bergon A., Holota H., Tamburini C., Garel M., Ollivier B., et al. (2014). Transcriptomics reveal several gene expression patterns in the piezophile Desulfovibrio hydrothermalis in response to hydrostatic pressure. PLoS One 9:e106831. doi: 10.1371/journal.pone.0106831, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

