Mutant huntingtin messenger RNA forms neuronal nuclear clusters in rodent and human brains
- PMID: 36458209
- PMCID: PMC9707646
- DOI: 10.1093/braincomms/fcac248
Mutant huntingtin messenger RNA forms neuronal nuclear clusters in rodent and human brains
Abstract
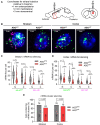
Mutant messenger RNA (mRNA) and protein contribute to the clinical manifestation of many repeat-associated neurological disorders, with the presence of nuclear RNA clusters being a common pathological feature. Yet, investigations into Huntington's disease-caused by a CAG repeat expansion in exon 1 of the huntingtin (HTT) gene-have primarily focused on toxic protein gain-of-function as the primary disease-causing feature. To date, mutant HTT mRNA has not been identified as an in vivo hallmark of Huntington's disease. Here, we report that, in two Huntington's disease mouse models (YAC128 and BACHD-97Q-ΔN17), mutant HTT mRNA is retained in the nucleus. Widespread formation of large mRNA clusters (∼0.6-5 µm3) occurred in 50-75% of striatal and cortical neurons. Cluster formation was independent of age and driven by expanded repeats. Clusters associate with chromosomal transcriptional sites and quantitatively co-localize with the aberrantly processed N-terminal exon 1-intron 1 mRNA isoform, HTT1a. HTT1a mRNA clusters are observed in a subset of neurons from human Huntington's disease post-mortem brain and are likely caused by somatic expansion of repeats. In YAC128 mice, clusters, but not individual HTT mRNA, are resistant to antisense oligonucleotide treatment. Our findings identify mutant HTT/HTT1a mRNA clustering as an early, robust molecular signature of Huntington's disease, providing in vivo evidence that Huntington's disease is a repeat expansion disease with mRNA involvement.
Keywords: Huntington’s disease; RNA fluorescence in situ hybridization; antisense oligonucleotides; mutant HTT mRNA; nuclear RNA clusters.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures







Similar articles
-
Alternative processing of human HTT mRNA with implications for Huntington's disease therapeutics.Brain. 2022 Dec 19;145(12):4409-4424. doi: 10.1093/brain/awac241. Brain. 2022. PMID: 35793238 Free PMC article.
-
Mutant huntingtin protein decreases with CAG repeat expansion: implications for therapeutics and bioassays.Brain Commun. 2024 Nov 15;6(6):fcae410. doi: 10.1093/braincomms/fcae410. eCollection 2024. Brain Commun. 2024. PMID: 39713241 Free PMC article.
-
Exon 1-targeting miRNA reduces the pathogenic exon 1 HTT protein in Huntington's disease models.Brain. 2024 Dec 3;147(12):4043-4055. doi: 10.1093/brain/awae266. Brain. 2024. PMID: 39155061 Free PMC article.
-
Huntingtin and its role in neuronal degeneration.Neuroscientist. 2004 Oct;10(5):467-75. doi: 10.1177/1073858404266777. Neuroscientist. 2004. PMID: 15359012 Review.
-
Huntington's disease mouse models: unraveling the pathology caused by CAG repeat expansion.Fac Rev. 2021 Oct 21;10:77. doi: 10.12703/r/10-77. eCollection 2021. Fac Rev. 2021. PMID: 34746930 Free PMC article. Review.
Cited by
-
Neuropathogenesis-on-chips for neurodegenerative diseases.Nat Commun. 2024 Mar 12;15(1):2219. doi: 10.1038/s41467-024-46554-8. Nat Commun. 2024. PMID: 38472255 Free PMC article. Review.
-
Systematic analysis of siRNA and mRNA features impacting fully chemically modified siRNA efficacy.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf479. doi: 10.1093/nar/gkaf479. Nucleic Acids Res. 2025. PMID: 40548938 Free PMC article.
-
Nuclear pore dysfunction and disease: a complex opportunity.Nucleus. 2024 Dec;15(1):2314297. doi: 10.1080/19491034.2024.2314297. Epub 2024 Feb 21. Nucleus. 2024. PMID: 38383349 Free PMC article. Review.
-
mRNA Nuclear Clustering Leads to a Difference in Mutant Huntingtin mRNA and Protein Silencing by siRNAs In Vivo.Nucleic Acid Ther. 2024 Aug;34(4):164-172. doi: 10.1089/nat.2024.0027. Epub 2024 Jul 18. Nucleic Acid Ther. 2024. PMID: 39023561 Free PMC article.
-
Blocking somatic repeat expansion and lowering huntingtin via RNA interference synergize to prevent Huntington's disease pathogenesis in mice.bioRxiv [Preprint]. 2025 Jun 25:2025.06.24.661398. doi: 10.1101/2025.06.24.661398. bioRxiv. 2025. PMID: 40667291 Free PMC article. Preprint.
References
-
- The Huntington’s Disease Collaborative Research Group . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72(6):971–983. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
