Transcriptome Analysis of Podoscypha petalodes Strain GGF6 Reveals the Diversity of Proteins Involved in Lignocellulose Degradation and Ligninolytic Function
- PMID: 36458217
- PMCID: PMC9705691
- DOI: 10.1007/s12088-022-01037-6
Transcriptome Analysis of Podoscypha petalodes Strain GGF6 Reveals the Diversity of Proteins Involved in Lignocellulose Degradation and Ligninolytic Function
Abstract
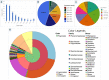
The present study reports transcriptomic profiling of a Basidiomycota fungus, Podoscypha petalodes strain GGF6 belonging to the family Podoscyphaceae, isolated from the North-Western Himalayan ranges in Himachal Pradesh, India. Podoscypha petalodes strain GGF6 possesses significant biotechnological potential as it has been reported for endocellulase, laccase, and other lignocellulolytic enzymes under submerged fermentation conditions. The present study attempts to enhance our knowledge of its lignocellulolytic potential as no previous omics-based analysis is available for this white-rot fungus. The transcriptomic analysis of P. petalodes GGF6 reveals the presence of 280 CAZy proteins. Furthermore, bioprospecting transcriptome signatures in the fungi revealed a diverse array of proteins associated with cellulose, hemicellulose, pectin, and lignin degradation. Interestingly, two copper-dependent lytic polysaccharide monooxygenases (AA14) and one pyrroloquinolinequinone-dependent oxidoreductase (AA12) were also identified, which are known to help in the lignocellulosic plant biomass degradation. Overall, this transcriptome profiling-based study provides deeper molecular-level insights into this Basidiomycota fungi, P. petalodes, for its potential application in diverse biotechnological applications, not only in the biofuel industry but also in the environmental biodegradation of recalcitrant molecules.
Supplementary information: The online version contains supplementary material available at 10.1007/s12088-022-01037-6.
Keywords: CAZyme; Fungal diversity; Laccase; Ligninolytic; Lignocellulose; Transcriptome.
© Association of Microbiologists of India 2022, Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare no conflict of interest to disclose.
Figures


References
-
- Mahajan R, Chandel S, Puniya AK, Goel G. Effect of pretreatments on cellulosic composition and morphology of pine needle for possible utilization as substrate for anaerobic digestion. Biomass Bioenergy. 2020;141:105705. doi: 10.1016/j.biombioe.2020.105705. - DOI
-
- Miyauchi S, Hage H, Drula E, Lesage-Meessen L, Berrin JG, Navarro D, Favel A, Chaduli D, Grisel S, Haon M, Piumi F, Levasseur A, Lomascolo A, Ahrendt S, Barry K, LaButti KM, Chevret D, Daum C, Mariette J, Klopp C, Cullen D, de Vries RP, Gathman AC, Hainaut M, Henrissat B, Hildén KS, Kües U, Lilly W, Lipzen A, Mäkelä MR, Martinez AT, Morel-Rouhier M, Morin E, Pangilinan J, Ram AFJ, Wösten HAB, Ruiz-Dueñas FJ, Riley R, Record E, Grigoriev IV, Rosso MN. Conservedwhite-rot enzymaticmechanismfor wood decay in the basidiomycota genus pycnoporus. DNA Res. 2020 doi: 10.1093/DNARES/DSAA011. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
