Mutation accumulation in mtDNA of cancers resembles mutagenesis in normal stem cells
- PMID: 36458259
- PMCID: PMC9707047
- DOI: 10.1016/j.isci.2022.105610
Mutation accumulation in mtDNA of cancers resembles mutagenesis in normal stem cells
Abstract
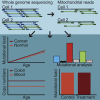
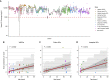
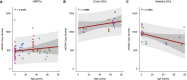
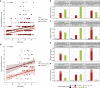
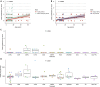
Mitochondria are small organelles that play an essential role in the energy production of eukaryotic cells. Defects in their genomes are associated with diseases, such as aging and cancer. Here, we analyzed the mitochondrial genomes of 532 whole-genome sequencing samples from cancers and normal clonally expanded single cells. We show that the mitochondria of normal cells accumulate mutations with age and that most of the mitochondrial mutations found in cancer are the result of healthy mutation accumulation. We also show that the normal HSPCs of patients with leukemia have an increased mitochondrial mutation load. Finally, we show that secondary pediatric cancers and chemotherapy treatments do not impact the mitochondrial mutation load and mtDNA copy numbers of most cells, suggesting that damage to the mitochondrial genome is not a major driver for carcinogenesis. Overall, these findings may contribute to our understanding of mitochondrial genomes and their role in cancer.
Keywords: Cancer; Genomics; Stem cells research.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
LinkOut - more resources
Full Text Sources

