Molecular evolution of SARS-CoV-2 from December 2019 to August 2022
- PMID: 36458547
- PMCID: PMC9877913
- DOI: 10.1002/jmv.28366
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022
Abstract
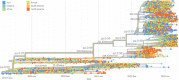
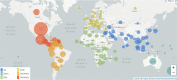
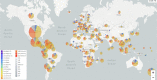
Severe acute respiratorysyndrome coronavirus-2 (SARS-CoV-2) pandemic spread rapidly and this scenario is concerning worldwide, presenting more than 590 million coronavirus disease 2019 cases and 6.4 million deaths. The emergence of novel lineages carrying several mutations in the spike protein has raised additional public health concerns worldwide during the pandemic. The present study review and summarizes the temporal spreading and molecular evolution of SARS-CoV-2 clades and variants worldwide. The evaluation of these data is important for understanding the evolutionary histories of SARSCoV-2 lineages, allowing us to identify the origins of each lineage of this virus responsible for one of the biggest pandemics in history. A total of 2897 SARS-CoV-2 whole-genome sequences with available information from the country and sampling date (December 2019 to August 2022), were obtained and were evaluated by Bayesian approach. The results demonstrated that the SARS-CoV-2 the time to the most recent common ancestor (tMRCA) in Asia was 2019-12-26 (highest posterior density 95% [HPD95%]: 2019-12-18; 2019-12-29), in Oceania 2020-01-24 (HPD95%: 2020-01-15; 2020-01-30), in Africa 2020-02-27 (HPD95%: 2020-02-21; 2020-03-04), in Europe 2020-02-27 (HPD95%: 2020-02-20; 2020-03-06), in North America 2020-03-12 (HPD95%: 2020-03-05; 2020-03-18), and in South America 2020-03-15 (HPD95%: 2020-03-09; 2020-03-28). Between December 2019 and June 2020, 11 clades were detected (20I [Alpha] and 19A, 19B, 20B, 20C, 20A, 20D, 20E [EU1], 20F, 20H [Beta]). From July to December 2020, 4 clades were identified (20J [Gamma, V3], 21 C [Epsilon], 21D [Eta], and 21G [Lambda]). Between January and June 2021, 3 clades of the Delta variant were detected (21A, 21I, and 21J). Between July and December 2021, two variants were detected, Delta (21A, 21I, and 21J) and Omicron (21K, 21L, 22B, and 22C). Between January and June 2022, the Delta (21I and 21J) and Omicron (21K, 21L, and 22A) variants were detected. Finally, between July and August 2022, 3 clades of Omicron were detected (22B, 22C, and 22D). Clade 19A was first detected in the SARS-CoV-2 pandemic (Wuhan strain) with origin in 2019-12-16 (HPD95%: 2019-12-15; 2019-12-25); 20I (Alpha) in 2020-11-24 (HPD95%: 2020-11-15; 2021-12-02); 20H (Beta) in 2020-11-25 (HPD95%: 2020-11-13; 2020-11-29); 20J (Gamma) was 2020-12-21 (HPD95%: 2020-11-05; 2021-01-15); 21A (Delta) in 2020-09-20 (HPD95%: 2020-05-17; 2021-02-03); 21J (Delta) in 2021-02-26 (2020-11-02; 2021-04-24); 21M (Omicron) in 2021-01-25 (HPD95%: 2020-09-16; 2021-08-08); 21K (Omicron) in 2021-07-30 (HPD95%: 2021-05-30; 2021-10-19); 21L (Omicron) in 2021-10-03 (HPD95%: 2021-04-16; 2021-12-23); 22B (Omicron) in 2022-01-25 (HPD95%: 2022-01-10; 2022-02-05); 21L in 2021-12-20 (HPD95%: 2021-05-16; 2021-12-31). Currently, the Omicron variant predominates worldwide, with the 21L clade branching into 3 (22A, 22B, and 22C). Phylogeographic data showed that Alpha variant originated in the United Kingdom, Beta in South Africa, Gamma in Brazil, Delta in India, Omicron in South Africa, Mu in Colombia, Epsilon in the United States of America, and Lambda in Peru. The COVID-19 pandemic has had a significant impact on global health worldwide and the present study provides an overview of the molecular evolution of SARS-CoV-2 lineage clades (from the Wuhan strain to the currently circulating lineages of the Omicron).
Keywords: Bayesian; COVID-19; SARS-CoV-2; molecular evolution.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19).2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 34033342 Free Books & Documents.
-
Genome sequence diversity of SARS-CoV-2 in Serbia: insights gained from a 3-year pandemic study.Front Microbiol. 2024 Feb 27;15:1332276. doi: 10.3389/fmicb.2024.1332276. eCollection 2024. Front Microbiol. 2024. PMID: 38476954 Free PMC article.
-
[Genomic Characterization of SARS-CoV-2 Isolates Obtained from Antalya, Türkiye].Mikrobiyol Bul. 2024 Oct;58(4):433-447. doi: 10.5578/mb.20249643. Mikrobiyol Bul. 2024. PMID: 39745215 Turkish.
-
Molecular evolution of the human monkeypox virus.J Med Virol. 2023 Feb;95(2):e28533. doi: 10.1002/jmv.28533. J Med Virol. 2023. PMID: 36708096 Review.
-
Unique mutations in SARS-CoV-2 Omicron subvariants' non-spike proteins: Potential impacts on viral pathogenesis and host immune evasion.Microb Pathog. 2022 Sep;170:105699. doi: 10.1016/j.micpath.2022.105699. Epub 2022 Aug 6. Microb Pathog. 2022. PMID: 35944840 Free PMC article. Review.
Cited by
-
Nucleic Acid Probes in Bio-Imaging and Diagnostics: Recent Advances in ODN-Based Fluorescent and Surface-Enhanced Raman Scattering Nanoparticle and Nanostructured Systems.Molecules. 2023 Apr 18;28(8):3561. doi: 10.3390/molecules28083561. Molecules. 2023. PMID: 37110795 Free PMC article. Review.
-
Positive Diagnosis of COVID-19 in an Integrated Teaching and Healthcare Service and Its Associated Factors.Healthcare (Basel). 2024 Jul 12;12(14):1395. doi: 10.3390/healthcare12141395. Healthcare (Basel). 2024. PMID: 39057538 Free PMC article.
-
Dynamic clade transitions and the influence of vaccine rollout on the spatiotemporal circulation of SARS-CoV-2 variants in São Paulo, Brazil.Res Sq [Preprint]. 2024 Jan 22:rs.3.rs-3788142. doi: 10.21203/rs.3.rs-3788142/v1. Res Sq. 2024. Update in: NPJ Vaccines. 2024 Aug 10;9(1):145. doi: 10.1038/s41541-024-00933-w. PMID: 38343798 Free PMC article. Updated. Preprint.
-
The emergence and successful elimination of SARS-CoV-2 dominant strains with increasing epidemic potential in Taiwan's 2021 outbreak.Heliyon. 2023 Nov 20;9(12):e22436. doi: 10.1016/j.heliyon.2023.e22436. eCollection 2023 Dec. Heliyon. 2023. PMID: 38107297 Free PMC article.
-
Impact of vaccination and non-pharmacological interventions on COVID-19: a review of simulation modeling studies in Asia.Front Public Health. 2023 Sep 25;11:1252719. doi: 10.3389/fpubh.2023.1252719. eCollection 2023. Front Public Health. 2023. PMID: 37818298 Free PMC article. Review.
References
-
- John Hopkins University of Medicine. Coronavirus Resource Center. 2022. Accessed September 23, 2022. https://coronavirus.jhu.edu/map.html
-
- WHO (World Health Organization) . Coronavirus (COVID‐19) dashboard. 2022. Accessed September 24, 2022.
-
- WHO (World Health Organization) . Weekly epidemiological update on COVID. 2022. Accessed September 23, 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on...
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

