Comparison of secondary attack rate and viable virus shedding between patients with SARS-CoV-2 Delta and Omicron variants: A prospective cohort study
- PMID: 36458559
- PMCID: PMC9877691
- DOI: 10.1002/jmv.28369
Comparison of secondary attack rate and viable virus shedding between patients with SARS-CoV-2 Delta and Omicron variants: A prospective cohort study
Erratum in
-
Erratum.J Med Virol. 2023 Feb;95(2):e28515. doi: 10.1002/jmv.28515. J Med Virol. 2023. PMID: 36832545 Free PMC article. No abstract available.
Abstract
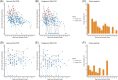
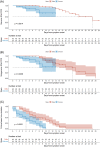
There are limited data comparing the transmission rates and kinetics of viable virus shedding of the Omicron variant to those of the Delta variant. We compared these rates in hospitalized patients infected with Delta and Omicron variants. We prospectively enrolled adult patients with COVID-19 admitted to a tertiary care hospital in South Korea between September 2021 and May 2022. Secondary attack rates were calculated by epidemiologic investigation, and daily saliva samples were collected to evaluate viral shedding kinetics. Genomic and subgenomic SARS-CoV-2 RNA was measured by PCR, and virus culture was performed from daily saliva samples. A total of 88 patients with COVID-19 who agreed to daily sampling and were interviewed, were included. Of the 88 patients, 48 (59%) were infected with Delta, and 34 (41%) with Omicron; a further 5 patients gave undetectable or inconclusive RNA PCR results and 1 was suspected of being coinfected with both variants. Omicron group had a higher secondary attack rate (31% [38/124] vs. 7% [34/456], p < 0.001). Survival analysis revealed that shorter viable virus shedding period was observed in Omicron variant compared with Delta variant (median 4, IQR [1-7], vs. 8.5 days, IQR [5-12 days], p < 0.001). Multivariable analysis revealed that moderate-to-critical disease severity (HR: 1.96), and immunocompromised status (HR: 2.17) were independent predictors of prolonged viral shedding, whereas completion of initial vaccine series or first booster-vaccinated status (HR: 0.49), and Omicron infection (HR: 0.44) were independently associated with shorter viable virus shedding. Patients with Omicron infections had higher transmission rates but shorter periods of transmissible virus shedding than those with Delta infections.
Keywords: COVID-19; infection dynamics; virus shedding.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- World Health Organization . Tracking SARS‐CoV‐2 variants. 2022. Accessed July 6, 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

