Spotting Local Environments in Self-Assembled Monolayer-Protected Gold Nanoparticles
- PMID: 36459668
- PMCID: PMC9798909
- DOI: 10.1021/acsnano.2c08467
Spotting Local Environments in Self-Assembled Monolayer-Protected Gold Nanoparticles
Abstract
Organic-inorganic (O-I) nanomaterials are versatile platforms for an incredible high number of applications, ranging from heterogeneous catalysis to molecular sensing, cell targeting, imaging, and cancer diagnosis and therapy, just to name a few. Much of their potential stems from the unique control of organic environments around inorganic sites within a single O-I nanomaterial, which allows for new properties that were inaccessible using purely organic or inorganic materials. Structural and mechanistic characterization plays a key role in understanding and rationally designing such hybrid nanoconstructs. Here, we introduce a general methodology to identify and classify local (supra)molecular environments in an archetypal class of O-I nanomaterials, i.e., self-assembled monolayer-protected gold nanoparticles (SAM-AuNPs). By using an atomistic machine-learning guided workflow based on the Smooth Overlap of Atomic Positions (SOAP) descriptor, we analyze a collection of chemically different SAM-AuNPs and detect and compare local environments in a way that is agnostic and automated, i.e., with no need of a priori information and minimal user intervention. In addition, the computational results coupled with experimental electron spin resonance measurements prove that is possible to have more than one local environment inside SAMs, being the thickness of the organic shell and solvation primary factors in the determining number and nature of multiple coexisting environments. These indications are extended to complex mixed hydrophilic-hydrophobic SAMs. This work demonstrates that it is possible to spot and compare local molecular environments in SAM-AuNPs exploiting atomistic machine-learning approaches, establishes ground rules to control them, and holds the potential for the rational design of O-I nanomaterials instructed from data.
Keywords: ESR; SOAP; fluorinated nanoparticles; machine learning; mixed monolayers; multiscale modeling; nanoconfinement.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





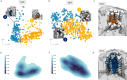
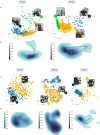
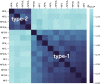

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

