Innate Immune Stimulation Using 3D Wireframe DNA Origami
- PMID: 36459697
- PMCID: PMC10144931
- DOI: 10.1021/acsnano.2c06275
Innate Immune Stimulation Using 3D Wireframe DNA Origami
Abstract
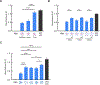
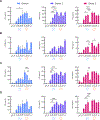
Three-dimensional wireframe DNA origami have programmable structural and sequence features that render them potentially suitable for prophylactic and therapeutic applications. However, their innate immunological properties, which stem from parameters including geometric shape and cytosine-phosphate-guanine dinucleotide (CpG) content, remain largely unknown. Here, we investigate the immunostimulatory properties of 3D wireframe DNA origami on the TLR9 pathway using both reporter cell lines and primary immune cells. Our results suggest that bare 3D polyhedral wireframe DNA origami induce minimal TLR9 activation despite the presence of numerous internal CpG dinucleotides. However, when displaying multivalent CpG-containing ssDNA oligos, wireframe DNA origami induce robust TLR9 pathway activation, along with enhancement of downstream immune response as evidenced by increases in Type I and Type III interferon (IFN) production in peripheral blood mononuclear cells. Further, we find that CpG copy number and spatial organization each contribute to the magnitude of TLR9 signaling and that NANP-attached CpGs do not require phosphorothioate stabilization to elicit signaling. These results suggest key design parameters for wireframe DNA origami that can be programmed to modulate immune pathway activation controllably for prophylactic and therapeutic applications.
Keywords: 3D wireframe DNA origami; CpG; TLR9; immunomodulation; immunostimulation; multivalency.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

