Discrete LAT condensates encode antigen information from single pMHC:TCR binding events
- PMID: 36460640
- PMCID: PMC9718779
- DOI: 10.1038/s41467-022-35093-9
Discrete LAT condensates encode antigen information from single pMHC:TCR binding events
Abstract
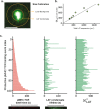
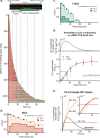
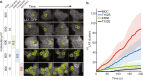
LAT assembly into a two-dimensional protein condensate is a prominent feature of antigen discrimination by T cells. Here, we use single-molecule imaging techniques to resolve the spatial position and temporal duration of each pMHC:TCR molecular binding event while simultaneously monitoring LAT condensation at the membrane. An individual binding event is sufficient to trigger a LAT condensate, which is self-limiting, and neither its size nor lifetime is correlated with the duration of the originating pMHC:TCR binding event. Only the probability of the LAT condensate forming is related to the pMHC:TCR binding dwell time. LAT condenses abruptly, but after an extended delay from the originating binding event. A LAT mutation that facilitates phosphorylation at the PLC-γ1 recruitment site shortens the delay time to LAT condensation and alters T cell antigen specificity. These results identify a function for the LAT protein condensation phase transition in setting antigen discrimination thresholds in T cells.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- François P, Altan-Bonnet G. The case for absolute ligand discrimination: modeling information processing and decision by immune T Cells. J. Stat. Phys. 2016;162:1130–1152. doi: 10.1007/s10955-015-1444-1. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

