An NKX-COUP-TFII morphogenetic code directs mucosal endothelial addressin expression
- PMID: 36460642
- PMCID: PMC9718832
- DOI: 10.1038/s41467-022-34991-2
An NKX-COUP-TFII morphogenetic code directs mucosal endothelial addressin expression
Abstract
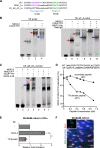
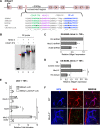
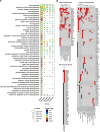
Immunoglobulin family and carbohydrate vascular addressins encoded by Madcam1 and St6gal1 control lymphocyte homing into intestinal tissues, regulating immunity and inflammation. The addressins are developmentally programmed to decorate endothelial cells lining gut post-capillary and high endothelial venules (HEV), providing a prototypical example of organ- and segment-specific endothelial specialization. We identify conserved NKX-COUP-TFII composite elements (NCCE) in regulatory regions of Madcam1 and St6gal1 that bind intestinal homeodomain protein NKX2-3 cooperatively with venous nuclear receptor COUP-TFII to activate transcription. The Madcam1 element also integrates repressive signals from arterial/capillary Notch effectors. Pan-endothelial COUP-TFII overexpression induces ectopic addressin expression in NKX2-3+ capillaries, while NKX2-3 deficiency abrogates expression by HEV. Phylogenetically conserved NCCE are enriched in genes involved in neuron migration and morphogenesis of the heart, kidney, pancreas and other organs. Our results define an NKX-COUP-TFII morphogenetic code that targets expression of mucosal vascular addressins.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Aird WC. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ. Res. 2007;100:174–190. doi: 10.1161/01.RES.0000255690.03436.ae. - DOI - PubMed

