The endocytic recycling compartment serves as a viral factory for hepatitis E virus
- PMID: 36460928
- PMCID: PMC9718719
- DOI: 10.1007/s00018-022-04646-y
The endocytic recycling compartment serves as a viral factory for hepatitis E virus
Abstract
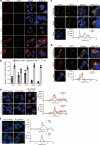
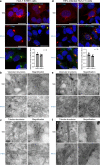
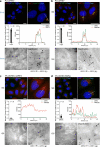
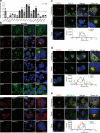
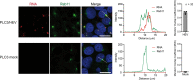
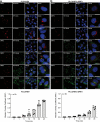
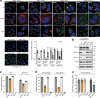
Although hepatitis E virus (HEV) is the major leading cause of enterically transmitted viral hepatitis worldwide, many gaps remain in the understanding of the HEV lifecycle. Notably, viral factories induced by HEV have not been documented yet, and it is currently unknown whether HEV infection leads to cellular membrane modeling as many positive-strand RNA viruses. HEV genome encodes the ORF1 replicase, the ORF2 capsid protein and the ORF3 protein involved in virion egress. Previously, we demonstrated that HEV produces different ORF2 isoforms including the virion-associated ORF2i form. Here, we generated monoclonal antibodies that specifically recognize the ORF2i form and antibodies that recognize the different ORF2 isoforms. One antibody, named P1H1 and targeting the ORF2i N-terminus, recognized delipidated HEV particles from cell culture and patient sera. Importantly, AlphaFold2 modeling demonstrated that the P1H1 epitope is exposed on HEV particles. Next, antibodies were used to probe viral factories in HEV-producing/infected cells. By confocal microscopy, we identified subcellular nugget-like structures enriched in ORF1, ORF2 and ORF3 proteins and viral RNA. Electron microscopy analyses revealed an unprecedented HEV-induced membrane network containing tubular and vesicular structures. We showed that these structures are dependent on ORF2i capsid protein assembly and ORF3 expression. An extensive colocalization study of viral proteins with subcellular markers, and silencing experiments demonstrated that these structures are derived from the endocytic recycling compartment (ERC) for which Rab11 is a central player. Hence, HEV hijacks the ERC and forms a membrane network of vesicular and tubular structures that might be the hallmark of HEV infection.
Keywords: AlphaFold2; Antibodies; Electron microscopy; Endocytic recycling compartment; Hepatitis E virus; Infectious particles; ORF2 capsid protein; Rab11; Viral factories.
© 2022. The Author(s).
Conflict of interest statement
Claire Montpellier, Jean Dubuisson and Laurence Cocquerel are coinventors of two patent applications on the use of antibodies having specificity for the ORF2i protein for HEV diagnostic purposes. Patents have been filed by Inserm Transfert. Julien Roux and Clarence Deffaud are employees of the BIOTEM company. The authors have no other financial or non-financial interests to disclose.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

