SARS-CoV-2 viral load and shedding kinetics
- PMID: 36460930
- PMCID: PMC9716513
- DOI: 10.1038/s41579-022-00822-w
SARS-CoV-2 viral load and shedding kinetics
Abstract
SARS-CoV-2 viral load and detection of infectious virus in the respiratory tract are the two key parameters for estimating infectiousness. As shedding of infectious virus is required for onward transmission, understanding shedding characteristics is relevant for public health interventions. Viral shedding is influenced by biological characteristics of the virus, host factors and pre-existing immunity (previous infection or vaccination) of the infected individual. Although the process of human-to-human transmission is multifactorial, viral load substantially contributed to human-to-human transmission, with higher viral load posing a greater risk for onward transmission. Emerging SARS-CoV-2 variants of concern have further complicated the picture of virus shedding. As underlying immunity in the population through previous infection, vaccination or a combination of both has rapidly increased on a global scale after almost 3 years of the pandemic, viral shedding patterns have become more distinct from those of ancestral SARS-CoV-2. Understanding the factors and mechanisms that influence infectious virus shedding and the period during which individuals infected with SARS-CoV-2 are contagious is crucial to guide public health measures and limit transmission. Furthermore, diagnostic tools to demonstrate the presence of infectious virus from routine diagnostic specimens are needed.
© 2022. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
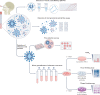
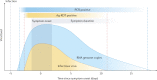
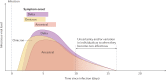
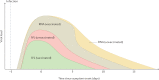
References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

