Anchoring super-enhancer-driven oncogenic lncRNAs for anti-tumor therapy in hepatocellular carcinoma
- PMID: 36461633
- PMCID: PMC10277835
- DOI: 10.1016/j.ymthe.2022.11.013
Anchoring super-enhancer-driven oncogenic lncRNAs for anti-tumor therapy in hepatocellular carcinoma
Abstract
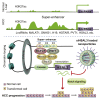
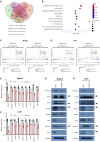
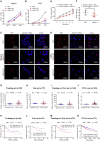
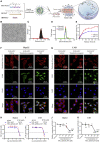
Super-enhancer (SE) plays a vital role in the determination of cell identity and fate. Up-regulated expression of coding genes is frequently associated with SE. However, the transcription dysregulation driven by SE, from the viewpoint of long non-coding RNA (lncRNA), remains unclear. Here, SE-associated lncRNAs in HCC are comprehensively outlined for the first time. This study integrally screens and identifies several novel SE-associated lncRNAs that are highly abundant and sensitive to JQ1. Especially, HSAL3 is identified as an uncharacterized SE-driven oncogenic lncRNA, which is activated by transcription factors HCFC1 and HSF1 via its super-enhancer. HSAL3 interference negatively regulates NOTCH signaling, implying the potential mechanism of its tumor-promoting role. The expression of HSAL3 is increased in HCC samples, and higher HSAL3 expression indicates an inferior overall survival of HCC patients. Furthermore, siHSAL3 loaded nanoparticles exert anti-tumor effect on HCC in vitro and in vivo. In conclusion, this is the first comprehensive survey of SE-associated lncRNAs in HCC. HSAL3 is a novel SE-driven oncogenic lncRNA, and siHSAL3 loaded nanoparticles are therapeutic candidates for HCC. This work sheds lights on the merit of anchoring SE-driven oncogenic lncRNAs for HCC treatment.
Keywords: HCC; HSAL3; enhancer; hepatocellular carcinoma; long non-coding RNA; nanoparticle; oncogene; super-enhancer; target therapy; transcription regulation.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. - PubMed
-
- Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. - PubMed
-
- Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2021;7:6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

