Pan-tumor landscape of fibroblast growth factor receptor 1-4 genomic alterations
- PMID: 36462464
- PMCID: PMC9832751
- DOI: 10.1016/j.esmoop.2022.100641
Pan-tumor landscape of fibroblast growth factor receptor 1-4 genomic alterations
Abstract
Background: Selective tyrosine kinase inhibitors targeting fibroblast growth factor receptor (FGFR) 1-4 genomic alterations are in development or have been approved for FGFR-altered cancers (e.g. bladder cancer and advanced intrahepatic cholangiocarcinoma). Understanding FGFR inhibitor-resistance mechanisms is increasingly relevant; we surveyed the pan-tumor landscape of FGFR1-4 genomic alterations [short variants (SVs), gene rearrangements (REs), and copy number alterations (CNAs)], including their association with tumor mutational burden (TMB) and the genomic comutational landscape.
Patients and methods: Comprehensive genomic profiling of 355 813 solid tumor clinical cases was performed using the FoundationOne and FoundationOne CDx assays (Foundation Medicine, Inc.) to identify genomic alterations in >300 cancer-associated genes and TMB (determined on ≤1.1 megabases of sequenced DNA).
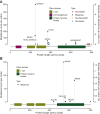
Results: FGFR1-4 SVs and REs occurred in 9603/355 813 (2.7%), and CNAs in 15 078/355 813 (4.2%) samples. Most common FGFR alterations for bladder cancer, intrahepatic cholangiocarcinoma, and glioma were FGFR3 SVs (1051/7739, 13.6%), FGFR2 REs (618/6641, 9.3%), and FGFR1 SVs (239/11 550, 2.1%), respectively. We found several, potentially clinically relevant, tumor-specific associations between FGFR1-4 genomic alterations and other genomic markers. FGFR3 SV-altered bladder cancers and FGFR1 SV-altered gliomas were significantly less likely to be TMB-high versus unaltered samples. FGFR3 SVs in bladder cancer significantly co-occurred with TERT and CDKN2A/B alterations; TP53 and RB1 alterations were mutually exclusive. In intrahepatic cholangiocarcinoma, FGFR2 REs significantly co-occurred with BAP1 alterations, whereas KRAS, TP53, IDH1, and ARID1A alterations were mutually exclusive. FGFR1 SVs in gliomas significantly co-occurred with H3-3A and PTPN11 alterations, but were mutually exclusive with TERT, EGFR, TP53, and CDKN2A/B alterations.
Conclusions: Overall, our hypothesis-generating findings may help to stratify patients in clinical trials and guide optimal targeted therapy in those with FGFR alterations.
Keywords: FGFR inhibitors; FGFR1-4; gene rearrangements/fusions; short variants; targeted therapy.
Copyright © 2022 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Disclosure KM is an employee of Foundation Medicine, Inc. and has stocks/shares in F. Hoffmann-La Roche Ltd. AN has been a steering committee member for Astellas, AstraZeneca, Bayer, Clovis Oncology, F. Hoffmann-La Roche Ltd, Janssen, and Merck; has received an institutional research grant from AstraZeneca, BMS, Gilead Sciences, Inc., Ipsen, and Merck; has a leadership role in the Global Society of Rare Genitourinary Tumors (GSRGT); is a coordinating principal investigator for Incyte; and is a local principal investigator for Pfizer. TCB was an employee of Incyte Corporation. OG is an employee of Foundation Medicine, Inc. and has stocks/shares in F. Hoffmann-La Roche Ltd. RG is an employee of Foundation Medicine, Inc. and has stocks/shares in F. Hoffmann-La Roche Ltd. MK has received institutional research funding for this study from F. Hoffmann-La Roche Ltd. JAL is an employee of and has stocks/shares in F. Hoffmann-La Roche Ltd. MM is an employee of Foundation Medicine, Inc. and has stocks/shares in F. Hoffmann-La Roche Ltd. HN is an employee of Foundation Medicine, Inc. and has stocks/shares in F. Hoffmann-La Roche Ltd. ARP is a consultant for Foundation Medicine, Inc. SR has received funding for an advisory board/research grant from F. Hoffmann-La Roche Ltd, AbbVie, Merck, Bayer, Incyte, and QED Therapeutics; and has received honoraria from Integrated DNA Technologies. SS is an employee of and has stocks/shares in F. Hoffmann-La Roche Ltd. IMS was an employee of Incyte Corporation. AV has performed a speaker, consultancy, and advisory role for AstraZeneca, Bayer, BMS, BTG, Daiichi Sankyo, Eisai, Eli Lilly and Company, F. Hoffmann-La Roche Ltd, GSK, Imaging Equipment (AAA), Incyte, Ipsen, Merck, MSD, Pierre Fabre, Sanofi, Servier, Sirtex, and Terumo. Data sharing Consented data that can be released are included in the article and its supplementary files. Patients were not consented for the release of underlying genomic sequence data. Academic researchers can gain access to Foundation Medicine data in this study by contacting the corresponding author and filling out a study review committee form. You and your institution will be required to execute a data transfer agreement. For further questions please reach out to Foundation Medicine, Cambridge, MA’s compliance department (compliance@foundationmedicine.com).
Figures




References
-
- Helsten T., Elkin S., Arthur E., Tomson B.N., Carter J., Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22:259–267. - PubMed
-
- Loriot Y., Necchi A., Park S.H., et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med. 2019;381:338–348. - PubMed
-
- Javle M.M., Roychowdhury S., Kelley R.K., et al. Final results from a phase II study of infigratinib (BGJ398), an FGFR-selective tyrosine kinase inhibitor, in patients with previously treated advanced cholangiocarcinoma harboring an FGFR2 gene fusion or rearrangement. J Clin Oncol. 2021;39:265. - PubMed
-
- Vogel A., Sahai V., Hollebecque A., et al. Pemigatinib for previously treated locally advanced or metastatic cholangiocarcinoma: final results from FIGHT-202. Ann Oncol. 2022;33 Abstract O-2.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

