Temperature instability of a mutation at a multidomain junction in Na,K-ATPase isoform ATP1A3 (p.Arg756His) produces a fever-induced neurological syndrome
- PMID: 36462665
- PMCID: PMC9860391
- DOI: 10.1016/j.jbc.2022.102758
Temperature instability of a mutation at a multidomain junction in Na,K-ATPase isoform ATP1A3 (p.Arg756His) produces a fever-induced neurological syndrome
Abstract
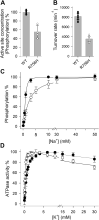
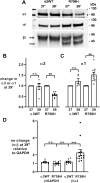
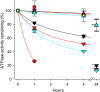
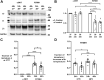
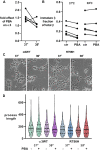
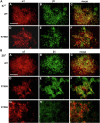
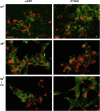
ATP1A3 encodes the α3 isoform of Na,K-ATPase. In the brain, it is expressed only in neurons. Human ATP1A3 mutations produce a wide spectrum of phenotypes, but particular syndromes are associated with unique substitutions. For arginine 756, at the junction of membrane and cytoplasmic domains, mutations produce encephalopathy during febrile infections. Here we tested the pathogenicity of p.Arg756His (R756H) in isogenic mammalian cells. R756H protein had sufficient transport activity to support cells when endogenous ATP1A1 was inhibited. It had half the turnover rate of wildtype, reduced affinity for Na+, and increased affinity for K+. There was modest endoplasmic reticulum retention during biosynthesis at 37 °C but little benefit from the folding drug phenylbutyrate (4-PBA), suggesting a tolerated level of misfolding. When cells were incubated at just 39 °C, however, α3 protein level dropped without loss of β subunit, paralleled by an increase of endogenous α1. Elevated temperature resulted in internalization of α3 from the surface along with some β subunit, accompanied by cytoplasmic redistribution of a marker of lysosomes and endosomes, lysosomal-associated membrane protein 1. After return to 37 °C, α3 protein levels recovered with cycloheximide-sensitive new protein synthesis. Heating in vitro showed activity loss at a rate 20- to 30-fold faster than wildtype, indicating a temperature-dependent destabilization of protein structure. Arg756 appears to confer thermal resistance as an anchor, forming hydrogen bonds among four linearly distant parts of the Na,K-ATPase structure. Taken together, our observations are consistent with fever-induced symptoms in patients.
Keywords: Na,K-ATPase; ataxia; cytopathology; fever; hypotonia; mutation pathogenicity; neurodegeneration; phenotype-genotype relationship; thermal inactivation.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest E. A., M. S. T-J., R. H., J. J-K., K. E. L., P. F., L. J. O., B. V., and K. J. S. declare no competing financial interests. A. B. serves the McKnight Brain Research Foundation as a trustee and is on the board of directors of the American Board of Psychiatry and Neurology and Care Directions.
Figures




References
-
- Lazarov E., Hillebrand M., Schroder S., Ternka K., Hofhuis J., Ohlenbusch A., et al. Comparative analysis of alternating hemiplegia of childhood and rapid-onset dystonia-parkinsonism ATP1A3 mutations reveals functional deficits, which do not correlate with disease severity. Neurobiol. Dis. 2020;143 - PubMed
-
- Toyoshima C., Nakasako M., Nomura H., Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. - PubMed
-
- Jorgensen P.L., Hakansson K.O., Karlish S.J.D. Structure and mechanism of Na,K-ATPase: Functional sites and their interactions. Ann. Rev. Physiol. 2003;65:817–849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

