The PPR-Domain Protein SOAR1 Regulates Salt Tolerance in Rice
- PMID: 36463341
- PMCID: PMC9719575
- DOI: 10.1186/s12284-022-00608-x
The PPR-Domain Protein SOAR1 Regulates Salt Tolerance in Rice
Abstract
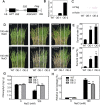
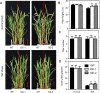
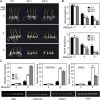
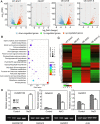
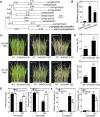
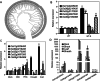
Previous studies in Arabidopsis reported that the PPR protein SOAR1 plays critical roles in plant response to salt stress. In this study, we reported that expression of the Arabidopsis SOAR1 (AtSOAR1) in rice significantly enhanced salt tolerance at seedling growth stage and promoted grain productivity under salt stress without affecting plant productivity under non-stressful conditions. The transgenic rice lines expressing AtSOAR1 exhibited increased ABA sensitivity in ABA-induced inhibition of seedling growth, and showed altered transcription and splicing of numerous genes associated with salt stress, which may explain salt tolerance of the transgenic plants. Further, we overexpressed the homologous gene of SOAR1 in rice, OsSOAR1, and showed that transgenic plants overexpressing OsSOAR1 enhanced salt tolerance at seedling growth stage. Five salt- and other abiotic stress-induced SOAR1-like PPRs were also identified. These data showed that the SOAR1-like PPR proteins are positively involved in plant response to salt stress and may be used for crop improvement in rice under salinity conditions through transgenic manipulation.
Keywords: Alternative splicing; Pentatricopeptide repeat (PPR) protein; Rice; SOAR1; Salt stress.
© 2022. The Author(s).
Conflict of interest statement
We declare that there are no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

