Learning continuous potentials from smFRET
- PMID: 36463404
- PMCID: PMC9892619
- DOI: 10.1016/j.bpj.2022.11.2947
Learning continuous potentials from smFRET
Abstract
Potential energy landscapes are useful models in describing events such as protein folding and binding. While single-molecule fluorescence resonance energy transfer (smFRET) experiments encode information on continuous potentials for the system probed, including rarely visited barriers between putative potential minima, this information is rarely decoded from the data. This is because existing analysis methods often model smFRET output assuming, from the onset, that the system probed evolves in a discretized state space to be analyzed within a hidden Markov model (HMM) paradigm. By contrast, here, we infer continuous potentials from smFRET data without discretely approximating the state space. We do so by operating within a Bayesian nonparametric paradigm by placing priors on the family of all possible potential curves. As our inference accounts for a number of required experimental features raising computational cost (such as incorporating discrete photon shot noise), the framework leverages a structured-kernel-interpolation Gaussian process prior to help curtail computational cost. We show that our structured-kernel-interpolation priors for potential energy reconstruction from smFRET analysis accurately infers the potential energy landscape from a smFRET binding experiment. We then illustrate advantages of structured-kernel-interpolation priors for potential energy reconstruction from smFRET over standard HMM approaches by providing information, such as barrier heights and friction coefficients, that is otherwise inaccessible to HMMs.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


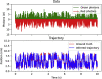


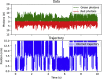

References
-
- Phillips R., Kondev J., et al. Garcia H. Garland Science; 2012. Physical Biology of the Cell.
-
- Schuler B., Lipman E.A., Eaton W.A. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. - PubMed
-
- Weistuch C., Pressé S. Spatiotemporal organization of catalysts driven by enhanced diffusion. J. Phys. Chem. B. 2018;122:5286–5290. - PubMed
-
- Makarov D.E. Barrier crossing dynamics from single-molecule measurements. J. Phys. Chem. B. 2021;125:2467–2476. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

