Automated enumeration of Eimeria oocysts in feces for rapid coccidiosis monitoring
- PMID: 36463777
- PMCID: PMC9719016
- DOI: 10.1016/j.psj.2022.102252
Automated enumeration of Eimeria oocysts in feces for rapid coccidiosis monitoring
Erratum in
-
Corrigendum to "Automated enumeration of Eimeria oocysts in feces for rapid coccidiosis monitoring" [Poultry Science, Volume 102, Issue 1, January 2023, 102252].Poult Sci. 2023 Mar;102(3):102446. doi: 10.1016/j.psj.2022.102446. Epub 2023 Jan 7. Poult Sci. 2023. PMID: 36624016 Free PMC article. No abstract available.
-
Corrigendum to "Automated enumeration of Eimeria oocysts in feces for rapid coccidiosis monitoring" [Poult. Sci. 102 (1) (2023) 102252].Poult Sci. 2023 Oct;102(10):102892. doi: 10.1016/j.psj.2023.102892. Epub 2023 Jul 19. Poult Sci. 2023. PMID: 37478621 Free PMC article. No abstract available.
Abstract
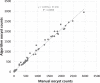
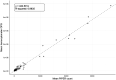
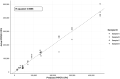
Coccidiosis represents a major driver in the economic performance of poultry operations, as coccidia control is expensive, and infections can result in increased feed conversion ratios, uneven growth rates, increased co-morbidities with pathogens such as Salmonella, and mortality within flocks. Shifts in broiler production to antibiotic-free strategies, increased attention on pre-harvest food safety, and growing incidence of anti-coccidial drug resistance has created a need for increased understanding of interventional efficacy and methods of coccidia control. Conventional methods to quantify coccidia oocysts in fecal samples involve manual microscopy processes that are time and labor intensive and subject to operator error, limiting their use as a diagnostic and monitoring tool in animal parasite control. To address the need for a high-throughput, robust, and reliable method to enumerate coccidia oocysts from poultry fecal samples, a novel diagnostic tool was developed. Utilizing the PIPER instrument and MagDrive technology, the diagnostic eliminates the requirement for extensive training and manual counting which currently limits the application of conventional microscopic methods of oocysts per gram (OPG) measurement. Automated microscopy to identify and count oocysts and report OPG simplifies analysis and removes potential sources of operator error. Morphometric analysis on identified oocysts allows for the oocyst counts to be separated into 3 size categories, which were shown to discriminate the 3 most common Eimeria species in commercial broilers, E. acervulina, E. tenella, and E. maxima. For 75% of the samples tested, the counts obtained by the PIPER and hemocytometer methods were within 2-fold of each other. Additionally, the PIPER method showed less variability than the hemocytometer counting method when OPG levels were below 100,000. By automated identification and counting of oocysts from 12 individual fecal samples in less than one hour, this tool could enable routine, noninvasive diagnostic monitoring of coccidia in poultry operations. This approach can generate large, uniform, and accurate data sets that create new opportunities for understanding the epidemiology and economics of coccidia infections and interventional efficacy.
Keywords: Eimeria enumeration; Eimeria oocyst; coccidia; coccidiosis monitoring.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Alowanou G.G., Adenile A., Akouedegni D., Bossou G.C.,, Zinsou A.C., Akakpo F.T., Kifouly G.-C.A., Rinaldi H.A., von Samson-Himmelstjerma L., Cringoli G., Hounzangbé-Adoté S. A comparison of Mini-FLOTAC and McMaster techniques in detecting gastrointestinal parasites in West Africa Dwarf sheep and goats and crossbreed rabbits. J. Appl. Anim. Res. 2021;49:30–38.
-
- Baba E., Fukata T., Arakawa A. Establishment and persistence of Salmonella typhimurium infection stimulated by Eimeria tenella in chickens. Res. Vet. Sci. 1982;33:95–98. - PubMed
-
- Blake D.P., Qin Z., Cai J., Smith A.L. Development and validation of real-time polymerase chain reaction assays specific to four species of Eimeria. Avian Pathol. 2008;37:89–94. - PubMed
-
- Blake D.P., Clark E.L., MacDonald S.E., Thenmozhi V., Kundu K., Garg R., Jatau I.D., Ayoade S., Kawahara F., Moftah A., Reid A.J. Population, genetic, and antigenic diversity of the apicomplexan Eimeria tenella and their relevance to vaccine development. Proc. Natl. Acad. Sci. 2015;112:E5343–E5350. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

