PERIOD 2 regulates low-dose radioprotection via PER2/pGSK3β/β-catenin/Per2 loop
- PMID: 36465103
- PMCID: PMC9708791
- DOI: 10.1016/j.isci.2022.105546
PERIOD 2 regulates low-dose radioprotection via PER2/pGSK3β/β-catenin/Per2 loop
Abstract
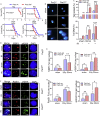
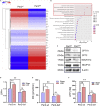
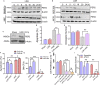
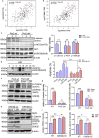
During evolution, humans are acclimatized to the stresses of natural radiation and circadian rhythmicity. Radiosensitivity of mammalian cells varies in the circadian period and adaptive radioprotection can be induced by pre-exposure to low-level radiation (LDR). It is unclear, however, if clock proteins participate in signaling LDR radioprotection. Herein, we demonstrate that radiosensitivity is increased in mice with the deficient Period 2 gene (Per2def) due to impaired DNA repair and mitochondrial function in progenitor bone marrow hematopoietic stem cells and monocytes. Per2 induction and radioprotection are also identified in LDR-treated Per2wt mouse cells and in human skin (HK18) and breast (MCF-10A) epithelial cells. LDR-boosted PER2 interacts with pGSK3β(S9) which activates β-catenin and the LEF/TCF mediated gene transcription including Per2 and genes involved in DNA repair and mitochondrial functions. This study demonstrates that PER2 plays an active role in LDR adaptive radioprotection via PER2/pGSK3β/β-catenin/Per2 loop, a potential target for protecting normal cells from radiation injury.
Keywords: Biological sciences; cell biology; molecular biology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Adams C., Blacker E., Burke W. Night shifts: circadian biology for public health. Nature. 2017;551:33. - PubMed
-
- Pekovic-Vaughan V., Gibbs J., Yoshitane H., Yang N., Pathiranage D., Guo B., Sagami A., Taguchi K., Bechtold D., Loudon A., et al. The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev. 2014;28:548–560. - PMC - PubMed
-
- Le A.N., Harton J., Desai H., Powers J., Zelley K., Bradbury A.R., Nathanson K.L., Shah P.D., Doucette A., Freedman G.M., et al. Frequency of radiation-induced malignancies post-adjuvant radiotherapy for breast cancer in patients with Li-Fraumeni syndrome. Breast Cancer Res. Treat. 2020;181:181–188. - PMC - PubMed
-
- Ishihara H., Tanaka I., Yakumaru H., Chikamori M., Ishihara F., Tanaka M., Ishiwata A., Kurematsu A., Satoh A., Ueda J., et al. Circadian transitions in radiation dose-dependent augmentation of mRNA levels for DNA damage-induced genes elicited by accurate real-time RT-PCR quantification. J. Radiat. Res. 2010;51:265–275. - PubMed

