Improved NGS-based detection of microsatellite instability using tumor-only data
- PMID: 36465367
- PMCID: PMC9714634
- DOI: 10.3389/fonc.2022.969238
Improved NGS-based detection of microsatellite instability using tumor-only data
Abstract
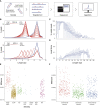
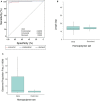
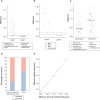
Microsatellite instability (MSI) is a molecular signature of mismatch repair deficiency (dMMR), a predictive marker of immune checkpoint inhibitor therapy response. Despite its recognized pan-cancer value, most methods only support detection of this signature in colorectal cancer. In addition to the tissue-specific differences that impact the sensitivity of MSI detection in other tissues, the performance of most methods is also affected by patient ethnicity, tumor content, and other sample-specific properties. These limitations are particularly important when only tumor samples are available and restrict the performance and adoption of MSI testing. Here we introduce MSIdetect, a novel solution for NGS-based MSI detection. MSIdetect models the impact of indel burden and tumor content on read coverage at a set of homopolymer regions that we found are minimally impacted by sample-specific factors. We validated MSIdetect in 139 Formalin-Fixed Paraffin-Embedded (FFPE) clinical samples from colorectal and endometrial cancer as well as other more challenging tumor types, such as glioma or sebaceous adenoma or carcinoma. Based on analysis of these samples, MSIdetect displays 100% specificity and 96.3% sensitivity. Limit of detection analysis supports that MSIdetect is sensitive even in samples with relatively low tumor content and limited microsatellite instability. Finally, the results obtained using MSIdetect in tumor-only data correlate well (R=0.988) with what is obtained using tumor-normal matched pairs, demonstrating that the solution addresses the challenges posed by MSI detection from tumor-only data. The accuracy of MSI detection by MSIdetect in different cancer types coupled with the flexibility afforded by NGS-based testing will support the adoption of MSI testing in the clinical setting and increase the number of patients identified that are likely to benefit from immune checkpoint inhibitor therapy.
Keywords: MSI; Microsatellite instability; Mismatch Repair deficiency; microsatellite; next-generating sequencing; pan-cancer; tumor-only sequencing.
Copyright © 2022 Marques, Ferraro-Peyret, Michaud, Song, Smith, Fabre, Willig, Wong, Xing, Chong, Brayer, Fenouil, Hervieu, Bancel, Devouassoux, Balme, Meyronet, Menu, Lopez and Xu.
Conflict of interest statement
AM, FM, LS, ES, GF, AW, MW, XX, CC, MB, PM, and ZX are SOPHiA GENETICS employees. CF-P reports sponsorship for meeting attendance from Roche and personal fees for advisory board work from Novartis, outside the submitted work. JL reports consulting for SOPHiA GENETICS and Decibio and personal fees for advisory board work and attendance to scientific meeting by Roche, Astra-Zeneca, BMS, Lilly and Nanostring. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

