Immunoinformatics design of multi-epitope peptide-based vaccine against Haemophilus influenzae strain using cell division protein
- PMID: 36465492
- PMCID: PMC9707196
- DOI: 10.1007/s13721-022-00395-x
Immunoinformatics design of multi-epitope peptide-based vaccine against Haemophilus influenzae strain using cell division protein
Abstract
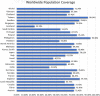
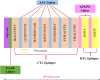
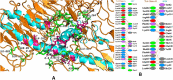
Haemophilus influenzae is a pathogen that causes invasive bacterial infections in humans. The highest prevalence lies in both young children and adults. Generally, there are no vaccines available that target all the strains of Haemophilus influenzae. Hence, the purpose of this research is to employ bioinformatics and immunoinformatics approaches to design a Multi-Epitope Vaccine candidate employing the pathogenic cell division protein FtsN that specifically combat all the Haemophilus influenzae strains. The current research focuses on developing subunit vaccine in contrast to vaccines generated from the entire pathogen. This will be accomplished by combining multiple bioinformatics and immunoinformatics approaches. As a result, prospective T cells (helper T lymphocyte and cytotoxic T lymphocytes) and B cells epitopes were investigated. The human leukocyte antigen allele having strong associations with the antigenic and overlapping epitopes were chosen, with 70% of the total coverage of the world population. To construct a linked vaccine design, multiple linkers were used. To increase the immunogenic profile, an adjuvant was linked using EAAAK linker. The final vaccine construct with 149 amino acids was obtained after adjuvants and linkers were added. The developed Multi-Epitope Vaccine has a high antigenicity as well as viable physiochemical features. The 3D conformation was modeled and undergoes refinement and validation using bioinformatics methods. Furthermore, protein-protein molecular docking analysis was performed to predict the effective binding poses of Multi-Epitope Vaccine with the Toll-like receptor 4 protein. Besides, vaccine underwent the codon translational optimization and computational cloning to verify the reliability and proper Multi-Epitope Vaccine expression. In addition, it is necessary to conduct experiments and research in the laboratory to demonstrate that the vaccine that has been developed is immunogenic and protective.
Keywords: Bioinformatics; Cell division protein; Epitope; Haemophilus influenzae; Linkers.
© The Author(s), under exclusive licence to Springer-Verlag GmbH Austria, part of Springer Nature 2022, Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Figures







References
LinkOut - more resources
Full Text Sources
Research Materials
