A meta-quantitative trait loci analysis identified consensus genomic regions and candidate genes associated with grain yield in rice
- PMID: 36466247
- PMCID: PMC9709451
- DOI: 10.3389/fpls.2022.1035851
A meta-quantitative trait loci analysis identified consensus genomic regions and candidate genes associated with grain yield in rice
Abstract
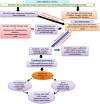
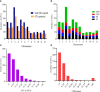
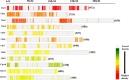
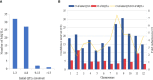
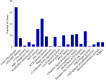
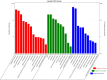
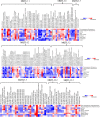
Improving grain yield potential in rice is an important step toward addressing global food security challenges. The meta-QTL analysis offers stable and robust QTLs irrespective of the genetic background of mapping populations and phenotype environment and effectively narrows confidence intervals (CI) for candidate gene (CG) mining and marker-assisted selection improvement. To achieve these aims, a comprehensive bibliographic search for grain yield traits (spikelet fertility, number of grains per panicle, panicles number per plant, and 1000-grain weight) QTLs was conducted, and 462 QTLs were retrieved from 47 independent QTL research published between 2002 and 2022. QTL projection was performed using a reference map with a cumulative length of 2,945.67 cM, and MQTL analysis was conducted on 313 QTLs. Consequently, a total of 62 MQTLs were identified with reduced mean CI (up to 3.40 fold) compared to the mean CI of original QTLs. However, 10 of these MQTLs harbored at least six of the initial QTLs from diverse genetic backgrounds and environments and were considered the most stable and robust MQTLs. Also, MQTLs were compared with GWAS studies and resulted in the identification of 16 common significant loci modulating the evaluated traits. Gene annotation, gene ontology (GO) enrichment, and RNA-seq analyses of chromosome regions of the stable MQTLs detected 52 potential CGs including those that have been cloned in previous studies. These genes encode proteins known to be involved in regulating grain yield including cytochrome P450, zinc fingers, MADs-box, AP2/ERF domain, F-box, ubiquitin ligase domain protein, homeobox domain, DEAD-box ATP domain, and U-box domain. This study provides the framework for molecular dissection of grain yield in rice. Moreover, the MQTLs and CGs identified could be useful for fine mapping, gene cloning, and marker-assisted selection to improve rice productivity.
Keywords: Genome-Wide Association Studies; Meta-QTL analysis; candidate genes; grain yield; marker-assisted selection; rice.
Copyright © 2022 Aloryi, Okpala, Amo, Bello, Akaba and Tian.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Acuña-Galindo M. A., Mason R. E., Subramanian N. K., Dirk B. H. (2015). Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 55, 477–492. doi: 10.2135/cropsci2013.11.0793 - DOI
-
- Ahmed E. Y. M., Zhang Y. P., Yu J. P., Rehman R. M. A., Zhang Z. Y., Zhang H. L., et al. (2016). Mapping of three QTLs for seed setting and analysis on the candidate gene for qSS-1 in rice (Oryza sativa l). J. Int. Agric. 15, 735–743. doi: 10.1016/S2095-3119(15)61299-0 - DOI
-
- Aquib A., Nafis S. (2021). Identifying meta-QTLs for stay-green in sorghum. bioRxiv. 4–11, 1–39. doi: 10.1101/2021.08.06.455413 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

