Comprehensive Multiomic Analysis Identified TUBA1C as a Potential Prognostic Biological Marker of Immune-Related Therapy in Pan-Cancer
- PMID: 36466547
- PMCID: PMC9713470
- DOI: 10.1155/2022/9493115
Comprehensive Multiomic Analysis Identified TUBA1C as a Potential Prognostic Biological Marker of Immune-Related Therapy in Pan-Cancer
Abstract
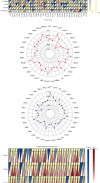
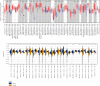
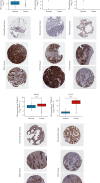
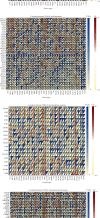
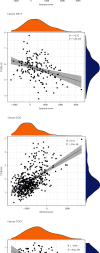
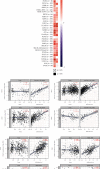
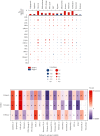
TUBA1C is correlated with an unfavourable prognosis and the infiltration of immune cells in several cancers. However, its function as a significant biomarker for the prognosis of immunotherapy in pan-cancer remains unclear. This study aims at assessing the role of TUBA1C in pan-cancer at multiple levels, including mutations, gene expression, methylation, m6A methylation, and immune cell infiltration levels. Data retrieved from major public databases, such as TCGA, GEO, GTEx, GSCA, CancerSEA, HPA, and RNAactDrugs, revealed that TUBA1C expression was high in 33 cancer types. Survival analysis revealed that TUBA1C was a poor prognostic factor for 12 tumour types, and mutations, CNVs, and methylation affected the prognosis of some cancer types. Furthermore, TUBA1C was found to be related to immune-related genes, immune cell infiltration, and the immune microenvironment. In addition, the sensitivity of 10 anticancer drugs was associated with high TUBA1C expression. Therefore, TUBA1C may serve as a viable prognostic biomarker for immunotherapy of pan-cancer.
Copyright © 2022 Yiming Zou et al.
Conflict of interest statement
The authors declare no conflicts of interest in this work.
Figures






Similar articles
-
TUBA1C is a Prognostic Marker in Low-grade Glioma and Correlates with Immune Cell Infiltration in the Tumor Microenvironment.Front Genet. 2021 Oct 14;12:759953. doi: 10.3389/fgene.2021.759953. eCollection 2021. Front Genet. 2021. PMID: 34721547 Free PMC article.
-
The oncogenic role of tubulin alpha-1c chain in human tumours.BMC Cancer. 2022 May 6;22(1):498. doi: 10.1186/s12885-022-09595-0. BMC Cancer. 2022. PMID: 35513790 Free PMC article.
-
TUBA1C is a potential new prognostic biomarker and promotes bladder urothelial carcinoma progression by regulating the cell cycle.BMC Cancer. 2023 Aug 1;23(1):716. doi: 10.1186/s12885-023-11209-2. BMC Cancer. 2023. PMID: 37528357 Free PMC article.
-
Cell Division Cycle-Associated Protein 3 (CDCA3) Is a Potential Biomarker for Clinical Prognosis and Immunotherapy in Pan-Cancer.Biomed Res Int. 2022 Aug 30;2022:4632453. doi: 10.1155/2022/4632453. eCollection 2022. Biomed Res Int. 2022. Retraction in: Biomed Res Int. 2023 Jun 21;2023:9870382. doi: 10.1155/2023/9870382. PMID: 36082153 Free PMC article. Retracted. Review.
-
A novel necroptosis-related gene index for predicting prognosis and a cold tumor immune microenvironment in stomach adenocarcinoma.Front Immunol. 2022 Oct 27;13:968165. doi: 10.3389/fimmu.2022.968165. eCollection 2022. Front Immunol. 2022. PMID: 36389725 Free PMC article. Review.
Cited by
-
The aggrephagy-related gene TUBA1B influences clinical outcomes in glioma patients by regulating the cell cycle.Front Oncol. 2025 Feb 28;15:1531465. doi: 10.3389/fonc.2025.1531465. eCollection 2025. Front Oncol. 2025. PMID: 40094001 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

