Circular RNA hsa_circ_0007990 as a blood biomarker for unruptured intracranial aneurysm with aneurysm wall enhancement
- PMID: 36466848
- PMCID: PMC9714537
- DOI: 10.3389/fimmu.2022.1061592
Circular RNA hsa_circ_0007990 as a blood biomarker for unruptured intracranial aneurysm with aneurysm wall enhancement
Abstract
Background: Circular RNAs (circRNAs) may involve the formation and rupture of intracranial aneurysms (IA). Inflammation plays a vital role in the development and progression of IA, which can be reflected by aneurysm wall enhancement (AWE) on high-resolution vessel wall magnetic resonance imaging (HR-VWI). This study aims to evaluate the role of circRNAs as the blood inflammatory biomarker for unruptured IA (UIA) patients with AWE on HR-VWI.
Methods: We analyzed the circRNA expression profiles in the peripheral blood samples among subjects from saccular UIA with AWE, UIA without AWE, and healthy controls by the circRNA microarray. The differential expression of hsa_circ_0007990 was assessed. We constructed the hsa_circ_0007990-microRNA-mRNA network and the regulatory axis of hub genes associated with the AWE in UIA.
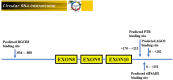
Results: Eighteen patients harboring saccular UIAs with HR VWI and five healthy controls were included. We found 412 differentially expressed circRNAs between UIA patients and healthy controls by circRNA microarray. Two hundred thirty-one circRNAs were significantly differentially expressed in UIA patients with AWE compared with those without AWE. Twelve upregulated circRNAs were associated with AWE of UIA, including hsa_circ_0007990, hsa_circ_0114507, hsa_circ_0020460, hsa_circ_0053944, hsa_circ_0000758, hsa_circ_0000034, hsa_circ_0009127, hsa_circ_0052793, hsa_circ_0000301 and hsa_circ_0000729. The expression of hsa_circ_0007990 was increased gradually in the healthy control, UIA without AWE, and UIA with AWE confirmed by RT-PCR (P<0.001). We predicted 4 RNA binding proteins (Ago2, DGCR8, EIF4A3, PTB) and period circadian regulator 1 as an encoding protein with hsa_circ_0007990. The hsa_circ_0007990-microRNA-mRNA network containing five microRNAs (miR-4717-5p, miR-1275, miR-150-3p, miR-18a-5p, miR-18b-5p), and 97 mRNAs was constructed. The five hub genes (hypoxia-inducible factor 1 subunit alpha, estrogen receptor 1, forkhead box O1, insulin-like growth factor 1, CREB binding protein) were involved in the inflammatory response.
Conclusion: Differentially expressed blood circRNAs associated with AWE on HR-VWI may be the novel inflammatory biomarkers for assessing UIA patients. The mechanism of hsa_circRNA_0007990 for UIA progression needs to investigate further.
Keywords: aneurysm wall enhancement; blood marker; circular RNA; inflammation; intracranial aneurysm; vessel wall imaging.
Copyright © 2022 Wu, Wu, Guo, Xiang, Chen, Qin and Shi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

