Origin, distribution, and function of three frequent coding polymorphisms in the gene for the human P2X7 ion channel
- PMID: 36467077
- PMCID: PMC9715982
- DOI: 10.3389/fphar.2022.1033135
Origin, distribution, and function of three frequent coding polymorphisms in the gene for the human P2X7 ion channel
Abstract
P2X7, an ion channel gated by extracellular ATP, is widely expressed on the plasma membrane of immune cells and plays important roles in inflammation and apoptosis. Several single nucleotide polymorphisms have been identified in the human P2RX7 gene. In contrast to other members of the P2X family, non-synonymous polymorphisms in P2X7 are common. Three of these occur at overall frequencies of more than 25% and affect residues in the extracellular "head"-domain of P2X7 (155 Y/H), its "lower body" (270 R/H), and its "tail" in the second transmembrane domain (348 T/A). Comparison of the P2X7 orthologues of human and other great apes indicates that the ancestral allele is Y-R-T (at 155-270-348). Interestingly, each single amino acid variant displays lower ATP-sensitivity than the ancestral allele. The originally published reference sequence of human P2X7, often referred to as "wildtype," differs from the ancestral allele at all three positions, i.e. H-H-A. The 1,000 Genome Project determined the sequences of both alleles of 2,500 human individuals, including roughly 500 persons from each of the five major continental regions. This rich resource shows that the ancestral alleles Y155, R270, and T348 occur in all analyzed human populations, albeit at strikingly different frequencies in various subpopulations (e.g., 25%-59% for Y155, 59%-77% for R270, and 13%-47% for T348). BLAST analyses of ancient human genome sequences uncovered several homozygous carriers of variant P2X7 alleles, possibly reflecting a high degree of inbreeding, e.g., H-R-T for a 50.000 year old Neanderthal, H-R-A for a 24.000 year old Siberian, and Y-R-A for a 7,000 year old mesolithic European. In contrast, most present-day individuals co-express two copies of P2X7 that differ in one or more amino acids at positions 155, 270, and 348. Our results improve the understanding of how P2X7 structure affects its function and suggest the importance of considering P2X7 variants of participants when designing clinical trials targeting P2X7.
Keywords: ATP; ATP-gated P2X ion channel; P2X7; adenosine triphosphate; ion channel; loss of function; purinergic receptor; single nucleotide polymorphism.
Copyright © 2022 Schäfer, Stähler, Pinto Espinoza, Danquah, Knop, Rissiek, Haag and Koch-Nolte.
Conflict of interest statement
WD and FK-N are coinventors on patent applications for P2X7-specific nanobodies. FK-N and FH obtain a share of the sales of antibody generated in their lab via MediGate GmbH, the technology transfer office and 100% subsidy of the University Hospital Hamburg-Eppendorf. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

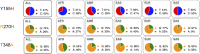

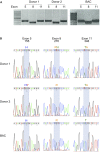


Similar articles
-
A His-155 to Tyr polymorphism confers gain-of-function to the human P2X7 receptor of human leukemic lymphocytes.J Immunol. 2005 Jul 1;175(1):82-9. doi: 10.4049/jimmunol.175.1.82. J Immunol. 2005. PMID: 15972634
-
A Thr357 to Ser polymorphism in homozygous and compound heterozygous subjects causes absent or reduced P2X7 function and impairs ATP-induced mycobacterial killing by macrophages.J Biol Chem. 2006 Jan 27;281(4):2079-86. doi: 10.1074/jbc.M507816200. Epub 2005 Nov 1. J Biol Chem. 2006. PMID: 16263709
-
Extracellular adenosine 5'-triphosphate induces a loss of CD23 from human dendritic cells via activation of P2X7 receptors.Int Immunol. 2002 Dec;14(12):1415-21. doi: 10.1093/intimm/dxf111. Int Immunol. 2002. PMID: 12456589
-
Human P2X7 receptors - Properties of single ATP-gated ion channels.Biochem Pharmacol. 2021 May;187:114307. doi: 10.1016/j.bcp.2020.114307. Epub 2020 Oct 29. Biochem Pharmacol. 2021. PMID: 33130127 Review.
-
The P2X7 Receptor.Adv Exp Med Biol. 2017;1051:17-53. doi: 10.1007/5584_2017_59. Adv Exp Med Biol. 2017. PMID: 28676924 Review.
Cited by
-
Selective regulation of IFN-γ and IL-4 co-producing unconventional T cells by purinergic signaling.J Exp Med. 2024 Dec 2;221(12):e20240354. doi: 10.1084/jem.20240354. Epub 2024 Nov 19. J Exp Med. 2024. PMID: 39560665 Free PMC article.
-
Trends in Research on the P2X7 Receptor: A Bibliometric and Visualization Analysis.J Inflamm Res. 2025 May 16;18:6349-6362. doi: 10.2147/JIR.S522380. eCollection 2025. J Inflamm Res. 2025. PMID: 40395554 Free PMC article. Review.
-
P2X7 Variants in Pathophysiology.Int J Mol Sci. 2024 Jun 18;25(12):6673. doi: 10.3390/ijms25126673. Int J Mol Sci. 2024. PMID: 38928378 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

