Identification of FDA-approved drugs against SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) through computational virtual screening
- PMID: 36467260
- PMCID: PMC9702953
- DOI: 10.1007/s11224-022-02072-1
Identification of FDA-approved drugs against SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) through computational virtual screening
Abstract
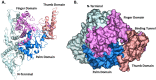
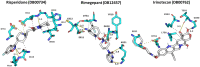
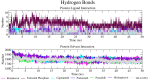
The SARS-CoV-2 coronavirus is responsible for the COVID-19 outbreak, which overwhelmed millions of people worldwide; hence, there is an urgency to identify appropriate antiviral drugs. This study focuses on screening compounds that inhibit RNA-dependent RNA-polymerase (RdRp) essential for RNA synthesis required for replication of positive-strand RNA viruses. Computational screening against RdRp using Food and Drug Administration (FDA)-approved drugs identified ten prominent compounds with binding energies of more than - 10.00 kcal/mol, each a potential inhibitor of RdRp. These compounds' binding energy is comparable to known RdRp inhibitors remdesivir (IC50 = 10.09 μM, SI = 4.96) and molnupiravir (EC50 = 0.67 - 2.66 µM) and 0.32-2.03 µM). Remdesivir and molnupiravir have been tested in clinical trial and remain authorized for emergency use in the treatment of COVID-19. In docking simulations, selected compounds are bound to the substrate-binding pocket of RdRp and showed hydrophobic and hydrogen bond interaction. For molecular dynamics simulation, capmatinib, pralsetinib, ponatinib, and tedizolid phosphate were selected from the initial ten candidate compounds. MD simulation indicated that these compounds are stable at 50-ns MD simulation when bound to RdRp protein. The screen hit compounds, remdesivir, molnupiravir, and GS-441524, are bound in the substrate binding pocket with good binding-free energy. As a consequence, capmatinib, pralsetinib, ponatinib, and tedizolid phosphate are potential new inhibitors of RdRp protein with potential of limiting COVID-19 infection by blocking RNA synthesis.
Supplementary information: The online version contains supplementary material available at 10.1007/s11224-022-02072-1.
Keywords: Docking; FDA; Molecular simulation; RNA-dependent RNA polymerase; SARS-CoV-2; Virtual screening.
© The Author(s) 2022.
Conflict of interest statement
Conflict of interestThe authors declare no competing interest.
Figures





Similar articles
-
Identification of FDA approved drugs against SARS-CoV-2 RNA dependent RNA polymerase (RdRp) and 3-chymotrypsin-like protease (3CLpro), drug repurposing approach.Biomed Pharmacother. 2021 Jun;138:111544. doi: 10.1016/j.biopha.2021.111544. Epub 2021 Mar 31. Biomed Pharmacother. 2021. PMID: 34311539 Free PMC article.
-
Revealing the Inhibition Mechanism of RNA-Dependent RNA Polymerase (RdRp) of SARS-CoV-2 by Remdesivir and Nucleotide Analogues: A Molecular Dynamics Simulation Study.J Phys Chem B. 2020 Nov 25;124(47):10641-10652. doi: 10.1021/acs.jpcb.0c06747. Epub 2020 Nov 15. J Phys Chem B. 2020. PMID: 33190493
-
Identification of RdRp inhibitors against SARS-CoV-2 through E-pharmacophore-based virtual screening, molecular docking and MD simulations approaches.Int J Biol Macromol. 2023 May 15;237:124169. doi: 10.1016/j.ijbiomac.2023.124169. Epub 2023 Mar 28. Int J Biol Macromol. 2023. PMID: 36990409 Free PMC article.
-
Investigating the potential of natural compounds as novel inhibitors of SARS-CoV-2 RdRP using computational approaches.Biotechnol Genet Eng Rev. 2024 Nov;40(3):1535-1555. doi: 10.1080/02648725.2023.2195240. Epub 2023 Mar 30. Biotechnol Genet Eng Rev. 2024. PMID: 36994810 Review.
-
A hypothesis on designing strategy of effective RdRp inhibitors for the treatment of SARS-CoV-2.3 Biotech. 2023 Jan;13(1):12. doi: 10.1007/s13205-022-03430-w. Epub 2022 Dec 16. 3 Biotech. 2023. PMID: 36532857 Free PMC article. Review.
Cited by
-
Molecular characterization of SARS-CoV-2 Omicron clade and clinical presentation in children.Sci Rep. 2024 Mar 4;14(1):5325. doi: 10.1038/s41598-024-55599-0. Sci Rep. 2024. PMID: 38438451 Free PMC article.
-
Novel analogues of a nonnucleoside SARS-CoV-2 RdRp inhibitor as potential antivirotics.Beilstein J Org Chem. 2024 May 6;20:1029-1036. doi: 10.3762/bjoc.20.91. eCollection 2024. Beilstein J Org Chem. 2024. PMID: 38746653 Free PMC article.
References
-
- World Health Organisation (2021) Weekly epidemiological update on COVID-19. WHO Emergency Situational Updates, Edition 47
-
- AlTakarli NS. China’s response to the COVID-19 outbreak: a model for epidemic preparedness and management. Dubai Medical Journal. 2020;3(2):44–49. doi: 10.1159/000508448. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
