Persistent dysregulation of genes in the development of endometriosis
- PMID: 36467354
- PMCID: PMC9708481
- DOI: 10.21037/atm-22-4806
Persistent dysregulation of genes in the development of endometriosis
Abstract
Background: Endometriosis is a chronic condition that affects women of child-bearing age. Since the etiology and pathogenesis of endometriosis have not been fully elucidated, it is important to investigate the mechanisms that lead to the deterioration of endometriosis.
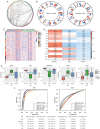
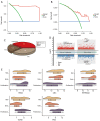
Methods: In this study, the transcriptome data of patients with normal, mild, and severe endometriosis were examined using the GSE51981 dataset obtained from the Gene Expression Omnibus database. Short Time Series Expression Miner (STEM) was used to screen the genes with continuous expression disorder in the development process, and the core genes were identified by constructing a protein-protein interaction network. The molecular mechanisms of endometriosis were examined using enrichment analysis. Finally, the transcription factors that regulate the core genes were predicted and the comprehensive mechanisms involved in the development of endometriosis were examined.
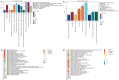
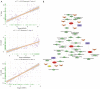
Results: A total of 3,472 differentially expressed genes were identified from the normal, mild, and severe endometriosis samples. These were allocated into 12 modules and HRAS, HSP90AA1, TGFB1, TP53, and UBC were selected as the core genes. Enrichment analysis showed that the genes in modules 6, 7, and 9 were significantly related to oxygen levels, metallic processes, and hormone levels, respectively. Transcription factor prediction analysis showed that TP53 regulates HRAS to participate in immune related signaling pathways. Drug prediction analysis identified 792 drugs that interact with the targeted core genes.
Conclusions: This study explored the molecular mechanisms involved in the development of endometriosis and identified potential biomarkers of endometriosis. This data may provide novel targets and research directions for the diagnosis and treatment of endometriosis.
Keywords: Endometriosis; Short Time Series Expression Miner (STEM); enrichment analysis; modules; protein-protein interaction network (PPI network).
2022 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4806/coif). The authors have no conflicts of interest to declare.
Figures


Similar articles
-
Identification of Potential Molecular Mechanism Related to Infertile Endometriosis.Front Vet Sci. 2022 Mar 28;9:845709. doi: 10.3389/fvets.2022.845709. eCollection 2022. Front Vet Sci. 2022. PMID: 35419445 Free PMC article.
-
Identification of molecular subtypes and immune infiltration in endometriosis: a novel bioinformatics analysis and In vitro validation.Front Immunol. 2023 Aug 18;14:1130738. doi: 10.3389/fimmu.2023.1130738. eCollection 2023. Front Immunol. 2023. PMID: 37662927 Free PMC article.
-
Bioinformatics Analysis Identifies Molecular Markers Regulating Development and Progression of Endometriosis and Potential Therapeutic Drugs.Front Genet. 2021 Aug 4;12:622683. doi: 10.3389/fgene.2021.622683. eCollection 2021. Front Genet. 2021. PMID: 34421979 Free PMC article.
-
Identification of potential differentially methylated gene-related biomarkers in endometriosis.Epigenomics. 2022 Oct;14(19):1157-1179. doi: 10.2217/epi-2022-0249. Epub 2022 Oct 31. Epigenomics. 2022. PMID: 36314280
-
Molecular mechanisms underlying endometriosis pathogenesis revealed by bioinformatics analysis of microarray data.Arch Gynecol Obstet. 2016 Apr;293(4):797-804. doi: 10.1007/s00404-015-3875-y. Epub 2015 Sep 9. Arch Gynecol Obstet. 2016. PMID: 26354330
Cited by
-
Evaluation of Proteasome and Immunoproteasome Levels in Plasma and Peritoneal Fluid in Patients with Endometriosis.Int J Mol Sci. 2023 Sep 21;24(18):14363. doi: 10.3390/ijms241814363. Int J Mol Sci. 2023. PMID: 37762666 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
