Estimation of Neutral Mutation Rates and Quantification of Somatic Variant Selection Using cancereffectsizeR
- PMID: 36469362
- PMCID: PMC9929515
- DOI: 10.1158/0008-5472.CAN-22-1508
Estimation of Neutral Mutation Rates and Quantification of Somatic Variant Selection Using cancereffectsizeR
Abstract
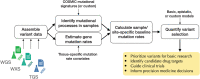
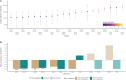
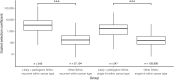
Somatic nucleotide mutations can contribute to cancer cell survival, proliferation, and pathogenesis. Although research has focused on identifying which mutations are "drivers" versus "passengers," quantifying the proliferative effects of specific variants within clinically relevant contexts could reveal novel aspects of cancer biology. To enable researchers to estimate these cancer effects, we developed cancereffectsizeR, an R package that organizes somatic variant data, facilitates mutational signature analysis, calculates site-specific mutation rates, and tests models of selection. Built-in models support effect estimation from single nucleotides to genes. Users can also estimate epistatic effects between paired sets of variants, or design and test custom models. The utility of cancer effect was validated by showing in a pan-cancer dataset that somatic variants classified as likely pathogenic or pathogenic in ClinVar exhibit substantially higher effects than most other variants. Indeed, cancer effect was a better predictor of pathogenic status than variant prevalence or functional impact scores. In addition, the application of this approach toward pairwise epistasis in lung adenocarcinoma showed that driver mutations in BRAF, EGFR, or KRAS typically reduce selection for alterations in the other two genes. Companion reference data packages support analyses using the hg19 or hg38 human genome builds, and a reference data builder enables use with any species or custom genome build with available genomic and transcriptomic data. A reference manual, tutorial, and public source code repository are available at https://townsend-lab-yale.github.io/cancereffectsizeR. Comprehensive estimation of cancer effects of somatic mutations can provide insights into oncogenic trajectories, with implications for cancer prognosis and treatment.
Significance: An R package provides streamlined, customizable estimation of underlying nucleotide mutation rates and of the oncogenic and epistatic effects of mutations in cancer cohorts.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures

References
-
- Prawira A, Pugh TJ, Stockley TL, Siu LL. Data resources for the identification and interpretation of actionable mutations by clinicians. Ann Oncol 2017;28:946–57. - PubMed
-
- Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R, et al. . Monitoring chronic lymphocytic leukemia progression by whole-genome sequencing reveals heterogeneous clonal evolution patterns. Blood 2012;120:4191–6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

