Regenerative failure of intrahepatic biliary cells in Alagille syndrome rescued by elevated Jagged/Notch/Sox9 signaling
- PMID: 36469766
- PMCID: PMC9897440
- DOI: 10.1073/pnas.2201097119
Regenerative failure of intrahepatic biliary cells in Alagille syndrome rescued by elevated Jagged/Notch/Sox9 signaling
Abstract
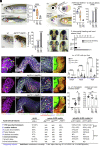
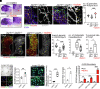
Despite the robust healing capacity of the liver, regenerative failure underlies numerous hepatic diseases, including the JAG1 haploinsufficient disorder, Alagille syndrome (ALGS). Cholestasis due to intrahepatic duct (IHD) paucity resolves in certain ALGS cases but fails in most with no clear mechanisms or therapeutic interventions. We find that modulating jag1b and jag2b allele dosage is sufficient to stratify these distinct outcomes, which can be either exacerbated or rescued with genetic manipulation of Notch signaling, demonstrating that perturbations of Jag/Notch signaling may be causal for the spectrum of ALGS liver severities. Although regenerating IHD cells proliferate, they remain clustered in mutants that fail to recover due to a blunted elevation of Notch signaling in the distal-most IHD cells. Increased Notch signaling is required for regenerating IHD cells to branch and segregate into the peripheral region of the growing liver, where biliary paucity is commonly observed in ALGS. Mosaic loss- and-gain-of-function analysis reveals Sox9b to be a key Notch transcriptional effector required cell autonomously to regulate these cellular dynamics during IHD regeneration. Treatment with a small-molecule putative Notch agonist stimulates Sox9 expression in ALGS patient fibroblasts and enhances hepatic sox9b expression, rescues IHD paucity and cholestasis, and increases survival in zebrafish mutants, thereby providing a proof-of-concept therapeutic avenue for this disorder.
Keywords: ALGS; Cholangiocytes; Jag1; Sox9; zebrafish.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Li L., et al. , Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 16, 243–251 (1997). - PubMed
-
- Oda T., et al. , Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 16, 235–242 (1997). - PubMed
-
- Emerick K. M., et al. , Features of Alagille syndrome in 92 patients: Frequency and relation to prognosis. Hepatology 29, 822–829 (1999). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

