FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2
- PMID: 36470304
- PMCID: PMC9977684
- DOI: 10.1038/s41586-022-05594-0
FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2
Abstract
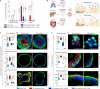
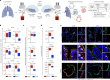
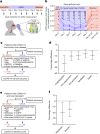
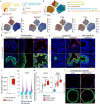
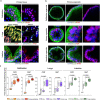
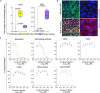
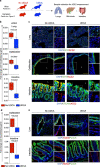
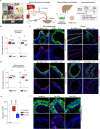
Preventing SARS-CoV-2 infection by modulating viral host receptors, such as angiotensin-converting enzyme 2 (ACE2)1, could represent a new chemoprophylactic approach for COVID-19 that complements vaccination2,3. However, the mechanisms that control the expression of ACE2 remain unclear. Here we show that the farnesoid X receptor (FXR) is a direct regulator of ACE2 transcription in several tissues affected by COVID-19, including the gastrointestinal and respiratory systems. We then use the over-the-counter compound z-guggulsterone and the off-patent drug ursodeoxycholic acid (UDCA) to reduce FXR signalling and downregulate ACE2 in human lung, cholangiocyte and intestinal organoids and in the corresponding tissues in mice and hamsters. We show that the UDCA-mediated downregulation of ACE2 reduces susceptibility to SARS-CoV-2 infection in vitro, in vivo and in human lungs and livers perfused ex situ. Furthermore, we reveal that UDCA reduces the expression of ACE2 in the nasal epithelium in humans. Finally, we identify a correlation between UDCA treatment and positive clinical outcomes after SARS-CoV-2 infection using retrospective registry data, and confirm these findings in an independent validation cohort of recipients of liver transplants. In conclusion, we show that FXR has a role in controlling ACE2 expression and provide evidence that modulation of this pathway could be beneficial for reducing SARS-CoV-2 infection, paving the way for future clinical trials.
© 2022. The Author(s).
Conflict of interest statement
F.S., L.V. and K.S.-P. are founders and shareholders of Bilitech. L.V. is a founder and shareholder of DEFINIGEN. The remaining authors declare no competing interests.
Figures






Comment in
-
Closing the door to SARS-CoV-2.Nat Rev Drug Discov. 2023 Feb;22(2):97. doi: 10.1038/d41573-022-00214-y. Nat Rev Drug Discov. 2023. PMID: 36517581 No abstract available.
-
Repurposing UDCA, an FXR Inhibitor, to Prevent SARS-Cov-2 Infection.Gastroenterology. 2023 May;164(6):1019-1020. doi: 10.1053/j.gastro.2023.01.014. Epub 2023 Jan 19. Gastroenterology. 2023. PMID: 36681157 Free PMC article. No abstract available.
-
FXR inhibition: an innovative prophylactic strategy against SARS-CoV-2 infection.Signal Transduct Target Ther. 2023 Mar 21;8(1):135. doi: 10.1038/s41392-023-01390-y. Signal Transduct Target Ther. 2023. PMID: 36944608 Free PMC article. No abstract available.
References
-
- World Health Organization. WHO Guidelines: Drugs to Prevent COVID-19https://www.who.int/publications/i/item/WHO-2019-nCoV-prophylaxes-2021-1 (WHO, 2021). - PubMed
-
- World Health Organization. Therapeutics and COVID-19: living guideline (version 9.3, 3 March 2022) (WHO, 2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

