Mapping the KRAS proteoform landscape in colorectal cancer identifies truncated KRAS4B that decreases MAPK signaling
- PMID: 36470426
- PMCID: PMC9808003
- DOI: 10.1016/j.jbc.2022.102768
Mapping the KRAS proteoform landscape in colorectal cancer identifies truncated KRAS4B that decreases MAPK signaling
Abstract
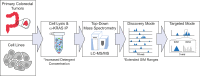
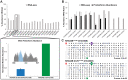
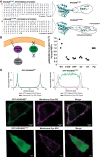
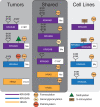
The KRAS gene is one of the most frequently mutated oncogenes in human cancer and gives rise to two isoforms, KRAS4A and KRAS4B. KRAS post-translational modifications (PTMs) have the potential to influence downstream signaling. However, the relationship between KRAS PTMs and oncogenic mutations remains unclear, and the extent of isoform-specific modification is unknown. Here, we present the first top-down proteomics study evaluating both KRAS4A and KRAS4B, resulting in 39 completely characterized proteoforms across colorectal cancer cell lines and primary tumor samples. We determined which KRAS PTMs are present, along with their relative abundance, and that proteoforms of KRAS4A versus KRAS4B are differentially modified. Moreover, we identified a subset of KRAS4B proteoforms lacking the C185 residue and associated C-terminal PTMs. By confocal microscopy, we confirmed that this truncated GFP-KRAS4BC185∗ proteoform is unable to associate with the plasma membrane, resulting in a decrease in mitogen-activated protein kinase signaling pathway activation. Collectively, our study provides a reference set of functionally distinct KRAS proteoforms and the colorectal cancer contexts in which they are present.
Keywords: KRAS; MAPK pathway; RAS protein; colorectal cancer; immunoprecipitation; post-translational modification; proteoform; top–down proteomics.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest N. L. K. is involved in commercialization of proteomics software. All other authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Barbacid M. Ras genes. Annu. Rev. Biochem. 1987;56:779–827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

