Genome-wide association study of REM sleep behavior disorder identifies polygenic risk and brain expression effects
- PMID: 36470867
- PMCID: PMC9722930
- DOI: 10.1038/s41467-022-34732-5
Genome-wide association study of REM sleep behavior disorder identifies polygenic risk and brain expression effects
Abstract
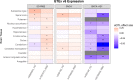
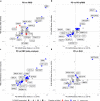
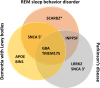
Rapid-eye movement (REM) sleep behavior disorder (RBD), enactment of dreams during REM sleep, is an early clinical symptom of alpha-synucleinopathies and defines a more severe subtype. The genetic background of RBD and its underlying mechanisms are not well understood. Here, we perform a genome-wide association study of RBD, identifying five RBD risk loci near SNCA, GBA, TMEM175, INPP5F, and SCARB2. Expression analyses highlight SNCA-AS1 and potentially SCARB2 differential expression in different brain regions in RBD, with SNCA-AS1 further supported by colocalization analyses. Polygenic risk score, pathway analysis, and genetic correlations provide further insights into RBD genetics, highlighting RBD as a unique alpha-synucleinopathy subpopulation that will allow future early intervention.
© 2022. The Author(s).
Conflict of interest statement
S.W.S. serves on the Scientific Advisory Council of the Lewy Body Dementia Association. S.W.S. is an editorial board member for JAMA Neurology and the Journal of Parkinson’s Disease. I.A. was previously consultant for Idorsia pharma, and UCB Pharma. A.D. served on the scientific advisory board for Eisai, UCB, Jazz Pharma, received research support from Jazz Pharma, Flamel Ireland, Canopy Growth, and honoraria from speaking engagements from Eisai and Sunovion. M.A.N.’s participation in this project was part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research, he also currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc as a data science fellow. Z.G.O. is supported by the Fonds de recherche du QuebecSante (FRQS) Chercheurs-boursiers award, and is a Parkinson’s Disease Canada New Investigator awardee. He received consultancy fees from Ono Therapeutics, Handl Therapeutics, Neuron23, Lysosomal Therapeutics Inc., Bial Biotech Inc., Deerfield, Lighthouse, and Idorsia, all unrelated to the current study. K.H., P.F., and the 23andMe Research Team are employed by and hold stock or stock options in 23andMe, Inc. The remaining authors declare no competing interests.
Figures




References
-
- Högl, B., Stefani, A. & Videnovic, A. J. N. R. N. Idiopathic REM sleep behaviour disorder and neurodegeneration—an update. Nat. Rev. Neurol. 14, 40 (2018). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

