Helical stability of the GnTV transmembrane domain impacts on SPPL3 dependent cleavage
- PMID: 36470941
- PMCID: PMC9722940
- DOI: 10.1038/s41598-022-24772-8
Helical stability of the GnTV transmembrane domain impacts on SPPL3 dependent cleavage
Abstract
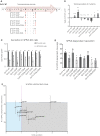
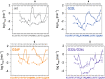
Signal-Peptide Peptidase Like-3 (SPPL3) is an intramembrane cleaving aspartyl protease that causes secretion of extracellular domains from type-II transmembrane proteins. Numerous Golgi-localized glycosidases and glucosyltransferases have been identified as physiological SPPL3 substrates. By SPPL3 dependent processing, glycan-transferring enzymes are deactivated inside the cell, as their active site-containing domain is cleaved and secreted. Thus, SPPL3 impacts on glycan patterns of many cellular and secreted proteins and can regulate protein glycosylation. However, the characteristics that make a substrate a favourable candidate for SPPL3-dependent cleavage remain unknown. To gain insights into substrate requirements, we investigated the function of a GxxxG motif located in the transmembrane domain of N-acetylglucosaminyltransferase V (GnTV), a well-known SPPL3 substrate. SPPL3-dependent secretion of the substrate's ectodomain was affected by mutations disrupting the GxxxG motif. Using deuterium/hydrogen exchange and NMR spectroscopy, we studied the effect of these mutations on the helix flexibility of the GnTV transmembrane domain and observed that increased flexibility facilitates SPPL3-dependent shedding and vice versa. This study provides first insights into the characteristics of SPPL3 substrates, combining molecular biology, biochemistry, and biophysical techniques and its results will provide the basis for better understanding the characteristics of SPPL3 substrates with implications for the substrates of other intramembrane proteases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

