Structure of SpoT reveals evolutionary tuning of catalysis via conformational constraint
- PMID: 36470996
- PMCID: PMC9974481
- DOI: 10.1038/s41589-022-01198-x
Structure of SpoT reveals evolutionary tuning of catalysis via conformational constraint
Abstract
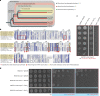
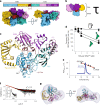
Stringent factors orchestrate bacterial cell reprogramming through increasing the level of the alarmones (p)ppGpp. In Beta- and Gammaproteobacteria, SpoT hydrolyzes (p)ppGpp to counteract the synthetase activity of RelA. However, structural information about how SpoT controls the levels of (p)ppGpp is missing. Here we present the crystal structure of the hydrolase-only SpoT from Acinetobacter baumannii and uncover the mechanism of intramolecular regulation of 'long'-stringent factors. In contrast to ribosome-associated Rel/RelA that adopt an elongated structure, SpoT assumes a compact τ-shaped structure in which the regulatory domains wrap around a Core subdomain that controls the conformational state of the enzyme. The Core is key to the specialization of long RelA-SpoT homologs toward either synthesis or hydrolysis: the short and structured Core of SpoT stabilizes the τ-state priming the hydrolase domain for (p)ppGpp hydrolysis, whereas the longer, more dynamic Core domain of RelA destabilizes the τ-state priming the monofunctional RelA for efficient (p)ppGpp synthesis.
© 2022. The Author(s).
Conflict of interest statement
A.G.-P. is cofounder and stockholder of Santero Therapeutics, a company that aims to develop inventions described in this paper. The other authors declare no competing interests.
Figures




References
-
- Anderson, B. W., Fung, D. K. & Wang, J. D. Regulatory themes and variations by the stress-signaling nucleotide alarmones (p)ppGpp in bacteria. Annu. Rev. Genet.23, 115–133 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

