Quantification of DNA methylation for carcinogenic risk estimation in patients with non-alcoholic steatohepatitis
- PMID: 36471401
- PMCID: PMC9724255
- DOI: 10.1186/s13148-022-01379-4
Quantification of DNA methylation for carcinogenic risk estimation in patients with non-alcoholic steatohepatitis
Abstract
Background: In recent years, non-alcoholic steatohepatitis (NASH) has become the main cause of hepatocellular carcinoma (HCC). As a means of improving the treatment of NASH-related HCCs based on early detection, this study investigated the feasibility of carcinogenic risk estimation in patients with NASH.
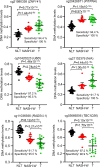
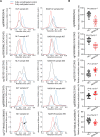
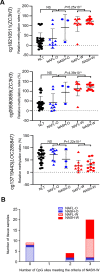
Results: Normal liver tissue (NLT), non-cancerous liver tissue showing histological findings compatible with non-alcoholic fatty liver from patients without HCC (NAFL-O), non-cancerous liver tissue showing NASH from patients without HCC (NASH-O), non-cancerous liver tissue showing non-alcoholic fatty liver from patients with HCC (NAFL-W), non-cancerous liver tissue showing NASH from patients with HCC (NASH-W) and NASH-related HCC were analyzed. An initial cohort of 171 tissue samples and a validation cohort of 55 tissue samples were used. Genome-wide DNA methylation screening using the Infinium HumanMethylation450 BeadChip and DNA methylation quantification using high-performance liquid chromatography (HPLC) with a newly developed anion-exchange column were performed. Based on the Infinium assay, 4050 CpG sites showed alterations of DNA methylation in NASH-W samples relative to NLT samples. Such alterations at the precancerous NASH stage were inherited by or strengthened in HCC samples. Receiver operating characteristic curve analysis identified 415 CpG sites discriminating NASH-W from NLT samples with area under the curve values of more than 0.95. Among them, we focused on 21 CpG sites showing more than 85% specificity, even for discrimination of NASH-W from NASH-O samples. The DNA methylation status of these 21 CpG sites was able to predict the coincidence of HCC independently from histopathological findings such as ballooning and fibrosis stage. The methylation status of 5 candidate marker CpG sites was assessed using a HPLC-based system, and for 3 of them sufficient sensitivity and specificity were successfully validated in the validation cohort. By combining these 3 CpG sites including the ZC3H3 gene, NAFL-W and NASH-W samples from which HCCs had already arisen were confirmed to show carcinogenic risk with 95% sensitivity in the validation cohort.
Conclusions: After a further prospective validation study using a larger cohort, carcinogenic risk estimation in liver biopsy specimens of patients with NASH may become clinically applicable using this HPLC-based system for quantification of DNA methylation.
Keywords: Carcinogenic risk estimation; DNA methylation; Hepatocellular carcinoma; High-performance liquid chromatography; Infinium assay; Non-alcoholic steatohepatitis.
© 2022. The Author(s).
Conflict of interest statement
None of the authors have any competing interests to disclose.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

