SARS-CoV-2 spike protein variant binding affinity to an angiotensin-converting enzyme 2 fusion glycoproteins
- PMID: 36472974
- PMCID: PMC9725131
- DOI: 10.1371/journal.pone.0278294
SARS-CoV-2 spike protein variant binding affinity to an angiotensin-converting enzyme 2 fusion glycoproteins
Abstract
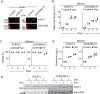
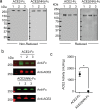
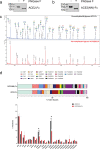
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of the Coronavirus disease 2019 (Covid-19) pandemic, continues to evolve and circulate globally. Current prophylactic and therapeutic countermeasures against Covid-19 infection include vaccines, small molecule drugs, and neutralizing monoclonal antibodies. SARS-CoV-2 infection is mainly mediated by the viral spike glycoprotein binding to angiotensin converting enzyme 2 (ACE2) on host cells for viral entry. As emerging mutations in the spike protein evade efficacy of spike-targeted countermeasures, a potential strategy to counter SARS-CoV-2 infection is to competitively block the spike protein from binding to the host ACE2 using a soluble recombinant fusion protein that contains a human ACE2 and an IgG1-Fc domain (ACE2-Fc). Here, we have established Chinese Hamster Ovary (CHO) cell lines that stably express ACE2-Fc proteins in which the ACE2 domain either has or has no catalytic activity. The fusion proteins were produced and purified to partially characterize physicochemical properties and spike protein binding. Our results demonstrate the ACE2-Fc fusion proteins are heavily N-glycosylated, sensitive to thermal stress, and actively bind to five spike protein variants (parental, alpha, beta, delta, and omicron) with different affinity. Our data demonstrates a proof-of-concept production strategy for ACE2-Fc fusion glycoproteins that can bind to different spike protein variants to support the manufacture of potential alternative countermeasures for emerging SARS-CoV-2 variants.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–44. Epub 2020/03/04. doi: 10.1038/s41564-020-0695-z ; PubMed Central PMCID: PMC7095448. - DOI - PMC - PubMed
-
- Organization WH. Tracking SARS-CoV-2 variants 2022. [updated 3/29/20224/4/2022]. Available from: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

