The evolutionary process of invasion in the fall armyworm (Spodoptera frugiperda)
- PMID: 36473923
- PMCID: PMC9727104
- DOI: 10.1038/s41598-022-25529-z
The evolutionary process of invasion in the fall armyworm (Spodoptera frugiperda)
Abstract
The fall armyworm (FAW; Spodoptera frugiperda) is one of the major agricultural pest insects. FAW is native to the Americas, and its invasion was first reported in West Africa in 2016. Then it quickly spread through Africa, Asia, and Oceania, becoming one of the main threats to corn production. We analyzed whole genome sequences of 177 FAW individuals from 12 locations on four continents to infer evolutionary processes of invasion. Principal component analysis from the TPI gene and whole genome sequences shows that invasive FAW populations originated from the corn strain. Ancestry coefficient and phylogenetic analyses from the nuclear genome indicate that invasive populations are derived from a single ancestry, distinct from native populations, while the mitochondrial phylogenetic tree supports the hypothesis of multiple introductions. Adaptive evolution specific to invasive populations was observed in detoxification, chemosensory, and digestion genes. We concluded that extant invasive FAW populations originated from the corn strain with potential contributions of adaptive evolution.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
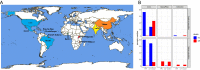

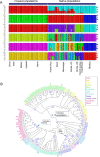


References
-
- Pimentel D, et al. Economic and environmental threats of alien plant, animal, and microbe invasions. Agric. Ecosyst. Environ. 2001;84:1–20. doi: 10.1016/S0167-8809(00)00178-X. - DOI
-
- Hulme PE. Trade, transport and trouble: managing invasive species pathways in an era of globalization. J. Appl. Ecol. 2009;46:10–18. doi: 10.1111/j.1365-2664.2008.01600.x. - DOI
-
- Bebber DP, Holmes T, Gurr SJ. The global spread of crop pests and pathogens. Glob. Ecol. Biogeogr. 2014;23:1398–1407. doi: 10.1111/geb.12214. - DOI
-
- McNeely JA. As the world gets smaller, the chances of invasion grow. Euphytica. 2006;148:5–15. doi: 10.1007/s10681-006-5937-5. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

