Preterm infants at low risk for early-onset sepsis differ in early fecal microbiome assembly
- PMID: 36474348
- PMCID: PMC9733690
- DOI: 10.1080/19490976.2022.2154091
Preterm infants at low risk for early-onset sepsis differ in early fecal microbiome assembly
Abstract
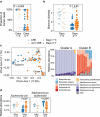
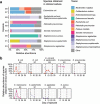
Antibiotics are administered near-universally to very low birth weight (VLBW) infants after birth for suspected early-onset sepsis (EOS). We previously identified a phenotypic group of VLBW infants, referred to as low-risk for EOS (LRE), whose risk of EOS is low enough to avoid routine antibiotic initiation. In this cohort study, we compared 18 such infants with 30 infants categorized as non-LRE to determine if the lower risk of pathogen transmission at birth is accompanied by differences in microbiome acquisition and development. We did shotgun metagenomic sequencing of 361 fecal samples obtained serially. LRE infants had a higher human-to-bacterial DNA ratio than non-LRE infants in fecal samples on days 1-3 after birth, confirming lower bacterial acquisition among LRE infants. The microbial diversity and composition in samples from days 4-7 differed between the groups with a predominance of Staphylococcus epidermidis in LRE infants and Enterobacteriaceae sp. in non-LRE infants. Compositional differences were congruent with the distribution of virulence factors and antibiotic resistant genes. After the first week, the overall composition was similar, but changes in relative abundance for several taxa with increasing age differed between groups. Of the nine late-onset bacteremia episodes, eight occurred in non-LRE infants. Species isolated from the blood culture was detected in the pre-antibiotic fecal samples of the infant for all episodes, though these species were also found in infants without bacteremia. In conclusion, LRE infants present a distinct pattern of microbiome development that is aligned with their low risk for EOS. Further investigation to determine the impact of these differences on later outcomes such as late-onset bacteremia is warranted.
Keywords: Preterm; low-risk; microbiome; sepsis; vertical transmission.
Figures



References
-
- Stoll BJ, Puopolo KM, Hansen NI, Sánchez PJ, Bell EF, Carlo WA, Cotten CM, D’Angio CT, Kazzi SNJ, Poindexter BB, et al. Early-onset neonatal sepsis 2015 to 2017, the rise of Escherichia coli, and the need for novel prevention strategies. JAMA Pediatr. 2020;174(7):e200593. doi: 10.1001/jamapediatrics.2020.0593. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
