Characterizing variability of electronic health record-driven phenotype definitions
- PMID: 36474423
- PMCID: PMC9933077
- DOI: 10.1093/jamia/ocac235
Characterizing variability of electronic health record-driven phenotype definitions
Abstract
Objective: The aim of this study was to analyze a publicly available sample of rule-based phenotype definitions to characterize and evaluate the variability of logical constructs used.
Materials and methods: A sample of 33 preexisting phenotype definitions used in research that are represented using Fast Healthcare Interoperability Resources and Clinical Quality Language (CQL) was analyzed using automated analysis of the computable representation of the CQL libraries.
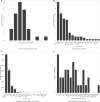
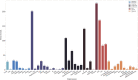
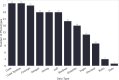
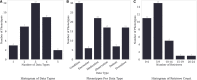
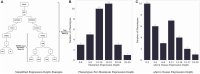
Results: Most of the phenotype definitions include narrative descriptions and flowcharts, while few provide pseudocode or executable artifacts. Most use 4 or fewer medical terminologies. The number of codes used ranges from 5 to 6865, and value sets from 1 to 19. We found that the most common expressions used were literal, data, and logical expressions. Aggregate and arithmetic expressions are the least common. Expression depth ranges from 4 to 27.
Discussion: Despite the range of conditions, we found that all of the phenotype definitions consisted of logical criteria, representing both clinical and operational logic, and tabular data, consisting of codes from standard terminologies and keywords for natural language processing. The total number and variety of expressions are low, which may be to simplify implementation, or authors may limit complexity due to data availability constraints.
Conclusions: The phenotype definitions analyzed show significant variation in specific logical, arithmetic, and other operators but are all composed of the same high-level components, namely tabular data and logical expressions. A standard representation for phenotype definitions should support these formats and be modular to support localization and shared logic.
Keywords: CQL; EHR-driven phenotyping; FHIR; cohort identification.
© The Author(s) 2022. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

References
Publication types
MeSH terms
Grants and funding
- U01 HG008676/HG/NHGRI NIH HHS/United States
- R01 GM139891/GM/NIGMS NIH HHS/United States
- U01 HG008657/HG/NHGRI NIH HHS/United States
- U01HG008657/HG/NHGRI NIH HHS/United States
- U01 HG008672/HG/NHGRI NIH HHS/United States
- U01 HG008679/HG/NHGRI NIH HHS/United States
- U01 HG008666/HG/NHGRI NIH HHS/United States
- U54 MD007593/MD/NIMHD NIH HHS/United States
- U01 HG008680/HG/NHGRI NIH HHS/United States
- U01 HG008673/HG/NHGRI NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- U01 HG008664/HG/NHGRI NIH HHS/United States
- U01 HG008701/HG/NHGRI NIH HHS/United States
- U01 HG008684/HG/NHGRI NIH HHS/United States
- U01 HG011169/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- P30 DK092949/DK/NIDDK NIH HHS/United States

